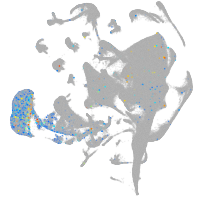
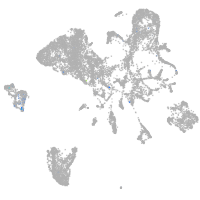
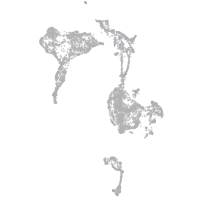
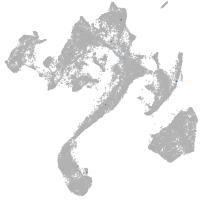
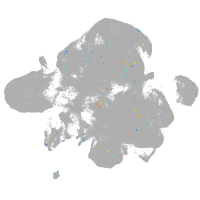
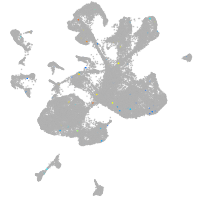
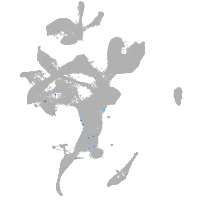
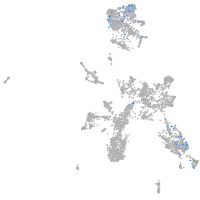
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| kcnj16 | 0.419 | gapdh | -0.058 |
| XLOC-018953 | 0.318 | gamt | -0.052 |
| glmnb | 0.295 | bhmt | -0.051 |
| LOC101884096 | 0.289 | pklr | -0.050 |
| epm2a | 0.287 | apoc1 | -0.048 |
| CABZ01066312.1 | 0.277 | fbp1b | -0.048 |
| dlx4a | 0.247 | gpx4a | -0.048 |
| LOC103910264 | 0.241 | pcbd1 | -0.046 |
| si:dkeyp-122a9.1 | 0.215 | gatm | -0.046 |
| si:dkey-4c15.13 | 0.214 | apoa1b | -0.046 |
| CR926130.1 | 0.209 | apoa4b.1 | -0.046 |
| XLOC-021899 | 0.208 | aqp12 | -0.045 |
| si:dkey-187a12.4 | 0.194 | apoc2 | -0.044 |
| rhcgb | 0.192 | afp4 | -0.044 |
| styk1a | 0.190 | agxta | -0.043 |
| arid6 | 0.188 | gpd1b | -0.043 |
| grapa | 0.186 | scp2a | -0.042 |
| krt1-19d | 0.183 | pnp4b | -0.041 |
| muc5f | 0.178 | apoa2 | -0.041 |
| anxa1b | 0.178 | tfa | -0.040 |
| pitpnm3 | 0.178 | abat | -0.040 |
| scel | 0.178 | kng1 | -0.040 |
| zgc:100868 | 0.178 | mat1a | -0.040 |
| lgals3bpa | 0.172 | hspe1 | -0.039 |
| spire1b | 0.172 | pgrmc1 | -0.039 |
| LOC100149931 | 0.172 | g6pca.2 | -0.039 |
| arg1 | 0.171 | serpina1l | -0.039 |
| LOC103910717 | 0.170 | vtnb | -0.038 |
| zgc:163083 | 0.170 | fgg | -0.038 |
| evpla | 0.169 | zgc:123103 | -0.038 |
| CR749763.6 | 0.168 | rbp2b | -0.038 |
| ponzr5 | 0.167 | fetub | -0.038 |
| slc2a11l | 0.164 | lgals2b | -0.038 |
| zgc:162184 | 0.164 | ces2 | -0.038 |
| mmrn2a | 0.164 | gcshb | -0.038 |