Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme 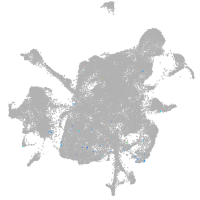 |
hematopoietic / vasculature  |
pronephros  |
neural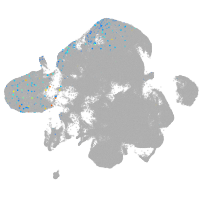 |
spinal cord / glia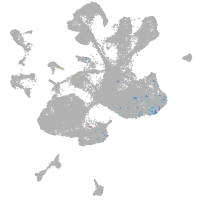 |
eye |
taste / olfactory |
otic / lateral line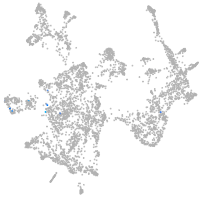 |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC110438435 | 0.126 | actc1b | -0.069 |
| cx43.4 | 0.095 | fabp3 | -0.067 |
| chrna4a | 0.089 | eef1da | -0.065 |
| cdx4 | 0.086 | ckma | -0.060 |
| LOC100330690 | 0.082 | ckmb | -0.059 |
| si:ch211-222l21.1 | 0.079 | atp2a1 | -0.059 |
| nucks1a | 0.078 | ttn.2 | -0.058 |
| anp32b | 0.078 | ak1 | -0.058 |
| hmga1a | 0.078 | aldoab | -0.058 |
| fgf8a | 0.078 | tmem38a | -0.057 |
| lig1 | 0.077 | pabpc4 | -0.057 |
| zgc:110425 | 0.077 | rpl37 | -0.056 |
| hmgb2a | 0.076 | mylpfa | -0.056 |
| nasp | 0.075 | eno1a | -0.056 |
| chaf1a | 0.075 | gamt | -0.055 |
| cirbpa | 0.075 | bhmt | -0.055 |
| slbp | 0.075 | acta1b | -0.055 |
| seta | 0.074 | tnnc2 | -0.054 |
| hes6 | 0.073 | eno3 | -0.054 |
| cbx3a | 0.073 | tpi1b | -0.054 |
| hnrnpaba | 0.073 | desma | -0.054 |
| si:ch73-281n10.2 | 0.073 | neb | -0.053 |
| apoeb | 0.072 | tpma | -0.053 |
| ilf3b | 0.072 | gapdh | -0.053 |
| smc1al | 0.072 | vdac3 | -0.052 |
| si:ch73-1a9.3 | 0.072 | srl | -0.052 |
| mcm3 | 0.072 | rps17 | -0.052 |
| rbbp4 | 0.072 | mybphb | -0.051 |
| si:ch211-114l13.1 | 0.072 | atp5if1b | -0.051 |
| anp32e | 0.072 | ldb3a | -0.051 |
| ptmab | 0.071 | pvalb1 | -0.051 |
| ncl | 0.071 | ldb3b | -0.051 |
| LOC110438285 | 0.071 | eif4a1b | -0.051 |
| BX649516.1 | 0.071 | mylz3 | -0.051 |
| marcksb | 0.071 | myl1 | -0.051 |