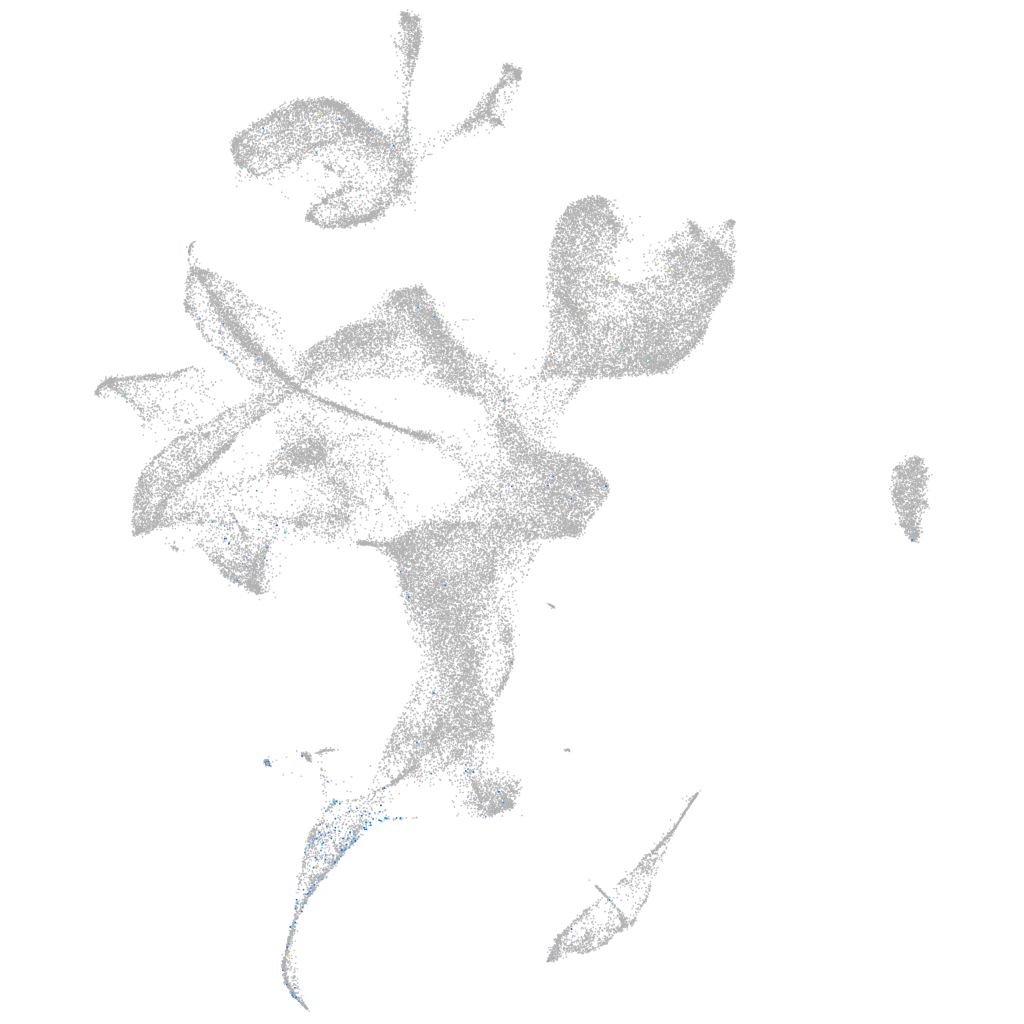
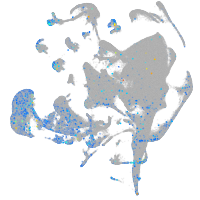
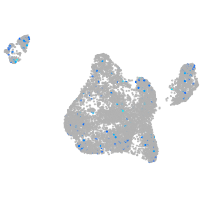
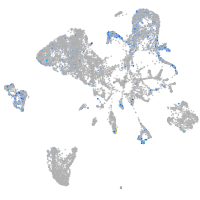
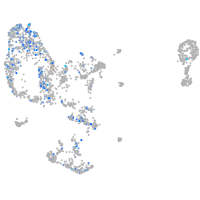
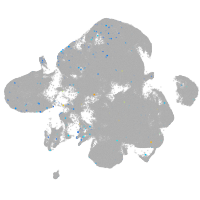
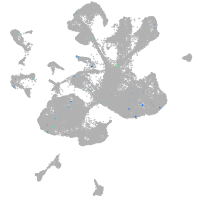
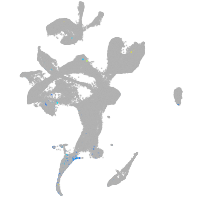
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CR855860.2 | 0.268 | ptmab | -0.064 |
| LOC110437895 | 0.261 | si:ch73-1a9.3 | -0.062 |
| slc45a2 | 0.176 | ptmaa | -0.046 |
| cngk | 0.170 | anp32e | -0.039 |
| tspan36 | 0.170 | fabp7a | -0.039 |
| gpr143 | 0.168 | hmgn6 | -0.039 |
| dct | 0.167 | gpm6aa | -0.039 |
| tyrp1b | 0.164 | si:ch211-137a8.4 | -0.038 |
| tyrp1a | 0.159 | tuba1c | -0.038 |
| oca2 | 0.159 | hmgb1a | -0.037 |
| pmela | 0.159 | cadm3 | -0.037 |
| agtrap | 0.156 | epb41a | -0.031 |
| mitfa | 0.155 | neurod4 | -0.030 |
| pmelb | 0.154 | hmgn2 | -0.030 |
| rab32a | 0.150 | zgc:153409 | -0.029 |
| tfec | 0.149 | snap25b | -0.028 |
| qdpra | 0.148 | rorb | -0.028 |
| rab38 | 0.147 | nova2 | -0.028 |
| FP085398.1 | 0.147 | gapdhs | -0.027 |
| pttg1ipb | 0.146 | foxg1b | -0.026 |
| cracr2ab | 0.146 | syt5b | -0.026 |
| tspan10 | 0.146 | neurod1 | -0.026 |
| bace2 | 0.142 | hes6 | -0.026 |
| gstp1 | 0.141 | vsx1 | -0.025 |
| zgc:110591 | 0.140 | crx | -0.025 |
| trpm1b | 0.138 | tmpob | -0.025 |
| cxcl18b | 0.137 | tuba1a | -0.025 |
| triobpa | 0.136 | sypb | -0.025 |
| slc24a5 | 0.135 | ncam1a | -0.025 |
| rnaseka | 0.135 | rs1a | -0.025 |
| tmem88b | 0.133 | six6b | -0.024 |
| pcbd1 | 0.131 | h2afva | -0.024 |
| sytl2b | 0.128 | marcksb | -0.023 |
| fabp11b | 0.128 | ckbb | -0.023 |
| rab27a | 0.127 | tox | -0.023 |