basic leucine zipper and W2 domains 1b
ZFIN 
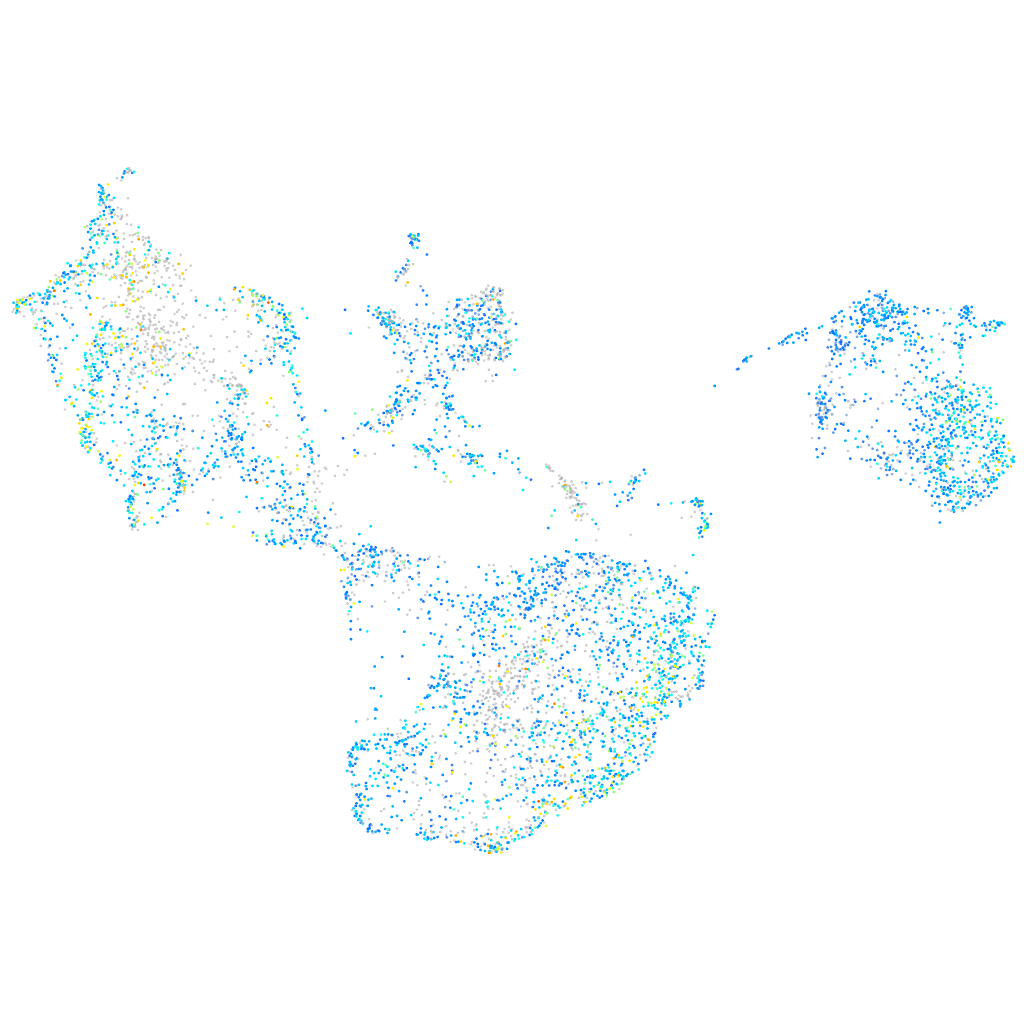
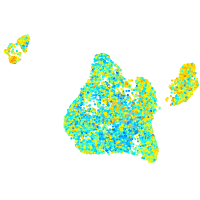
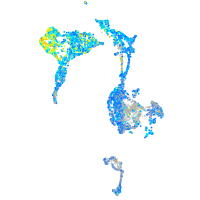
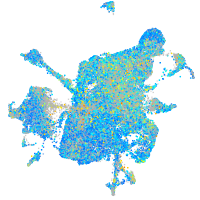
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| higd1a | 0.325 | NC-002333.4 | -0.146 |
| ddx5 | 0.320 | hbae3 | -0.122 |
| prelid3b | 0.306 | elavl3 | -0.120 |
| sdc4 | 0.300 | tuba1c | -0.119 |
| mcl1a | 0.299 | COX3 | -0.117 |
| zgc:162730 | 0.292 | NC-002333.17 | -0.113 |
| chac1 | 0.291 | rps29 | -0.111 |
| btg2 | 0.290 | cspg5a | -0.111 |
| cebpb | 0.282 | hbae1.1 | -0.110 |
| her9 | 0.281 | zgc:158463 | -0.109 |
| XLOC-044191 | 0.276 | hbbe1.3 | -0.107 |
| pmaip1 | 0.275 | stx1b | -0.105 |
| junba | 0.274 | stmn1b | -0.105 |
| xbp1 | 0.273 | gpm6ab | -0.104 |
| her6 | 0.266 | rpl38 | -0.104 |
| angptl4 | 0.264 | elavl4 | -0.100 |
| tob1a | 0.261 | gng3 | -0.099 |
| tob1b | 0.260 | gpm6aa | -0.099 |
| ubb | 0.260 | sncb | -0.098 |
| nfkbiab | 0.254 | stxbp1a | -0.098 |
| trib3 | 0.249 | si:ch73-1a9.3 | -0.097 |
| sat1a.2 | 0.248 | tuba1a | -0.097 |
| si:dkey-112e7.2 | 0.248 | rtn1a | -0.095 |
| hnrnpa0l | 0.245 | tmsb | -0.095 |
| midn | 0.244 | nova2 | -0.090 |
| fosb | 0.240 | mt-nd3 | -0.088 |
| slc25a5 | 0.240 | myt1la | -0.087 |
| ier5 | 0.240 | cnrip1a | -0.087 |
| cebpd | 0.238 | ckbb | -0.087 |
| dusp2 | 0.238 | hbbe1.1 | -0.085 |
| ier2a | 0.236 | rtn1b | -0.085 |
| nr4a1 | 0.236 | sv2a | -0.084 |
| ier2b | 0.235 | LOC110439372 | -0.084 |
| tdh | 0.235 | gap43 | -0.083 |
| fosab | 0.230 | ank2b | -0.082 |