"bloodthirsty-related gene family, member 31"
ZFIN 
Other cell groups


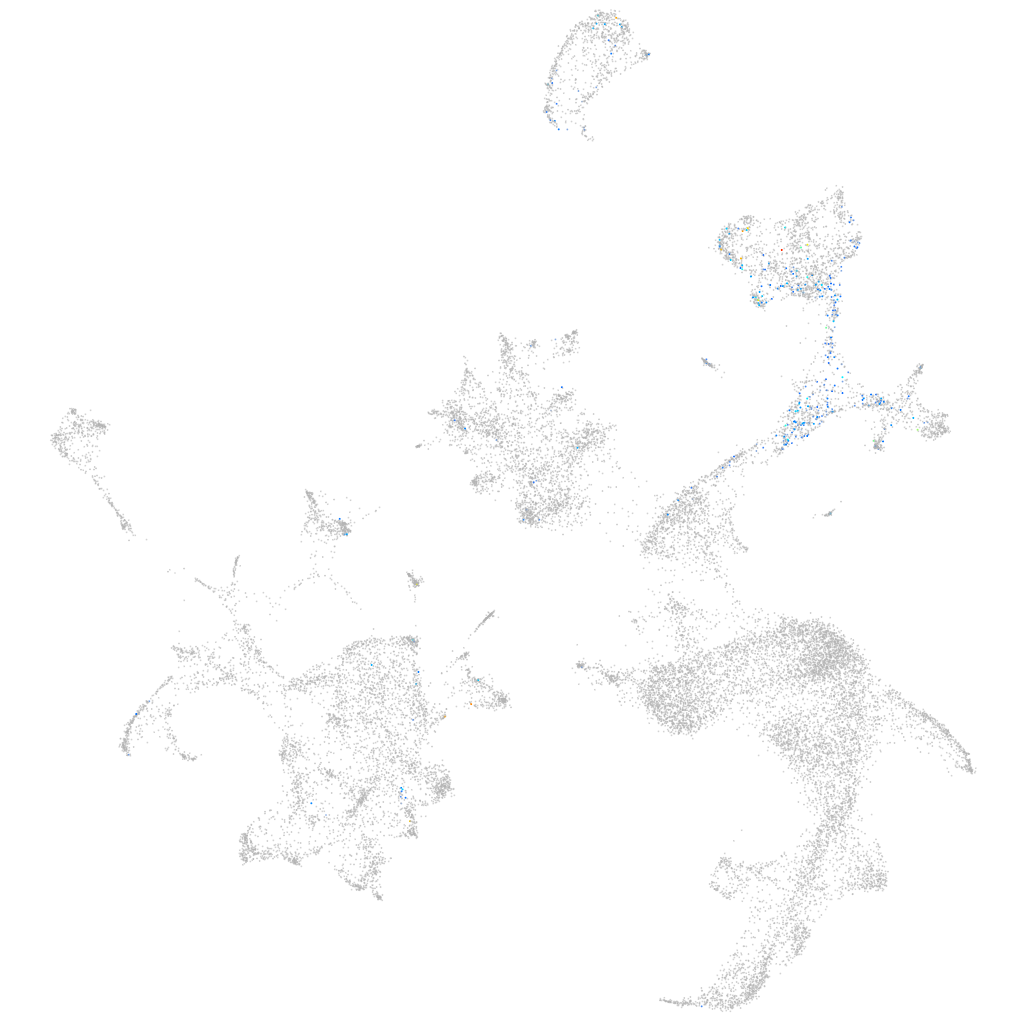
all cells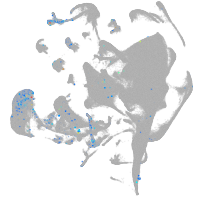 |
blastomeres |
PGCs |
endoderm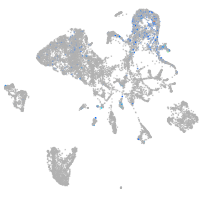 |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 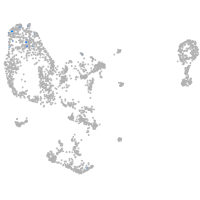 |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin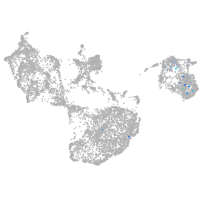 |
ionocytes / mucous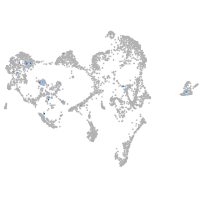 |
pigment cells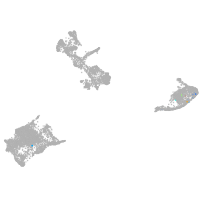 |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CU499330.1 | 0.168 | hbae3 | -0.097 |
| laptm5 | 0.155 | hbbe1.3 | -0.096 |
| cebpb | 0.155 | hbae1.3 | -0.095 |
| cd83 | 0.153 | hbae1.1 | -0.093 |
| FERMT3 (1 of many) | 0.145 | hbbe2 | -0.093 |
| mir183 | 0.144 | hbbe1.1 | -0.092 |
| coro1a | 0.140 | cahz | -0.088 |
| spi1a | 0.138 | hbbe1.2 | -0.084 |
| zgc:123297 | 0.138 | hemgn | -0.082 |
| cxcr3.2 | 0.137 | fth1a | -0.080 |
| pycard | 0.136 | blvrb | -0.077 |
| ctss2.2 | 0.136 | alas2 | -0.076 |
| XLOC-041870 | 0.136 | si:ch211-250g4.3 | -0.075 |
| arpc1b | 0.133 | zgc:56095 | -0.075 |
| irgf1 | 0.131 | creg1 | -0.074 |
| arhgdig | 0.131 | slc4a1a | -0.072 |
| ccr9a | 0.129 | nt5c2l1 | -0.070 |
| rgs13 | 0.129 | tspo | -0.069 |
| psma6l | 0.128 | zgc:163057 | -0.067 |
| ctsl.1 | 0.128 | epb41b | -0.067 |
| pfn1 | 0.128 | si:ch211-207c6.2 | -0.065 |
| RF00093 | 0.127 | tmod4 | -0.061 |
| cyba | 0.125 | aqp1a.1 | -0.060 |
| fcer1gl | 0.124 | nmt1b | -0.060 |
| nr4a1 | 0.123 | histh1l | -0.058 |
| lgals3bpb | 0.123 | plac8l1 | -0.058 |
| si:ch211-214p16.1 | 0.122 | hbae5 | -0.055 |
| grap2a | 0.121 | si:ch211-227m13.1 | -0.055 |
| cand2 | 0.121 | sptb | -0.054 |
| rac2 | 0.121 | hbbe3 | -0.052 |
| wasa | 0.121 | uros | -0.052 |
| si:dkey-27i16.2 | 0.119 | tfr1a | -0.051 |
| si:dkey-53k12.2 | 0.119 | add2 | -0.050 |
| eno3 | 0.118 | rfesd | -0.050 |
| XLOC-025423 | 0.118 | dmtn | -0.049 |