"bloodthirsty-related gene family, member 20"
ZFIN 
Other cell groups


all cells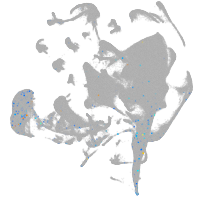 |
blastomeres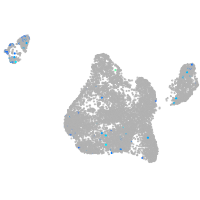 |
PGCs |
endoderm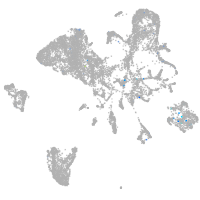 |
axial mesoderm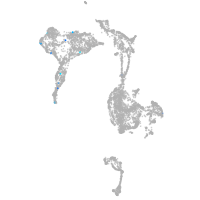 |
muscle 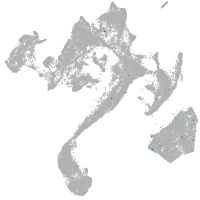 |
mural cells / non-skeletal muscle 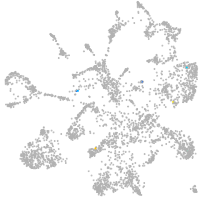 |
mesenchyme  |
hematopoietic / vasculature 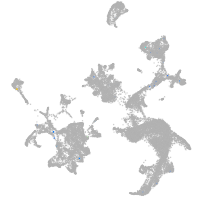 |
pronephros 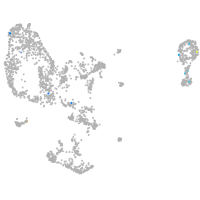 |
neural |
spinal cord / glia |
eye |
taste / olfactory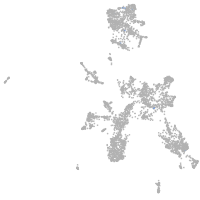 |
otic / lateral line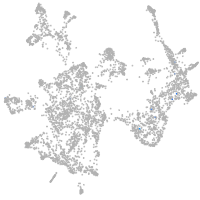 |
epidermis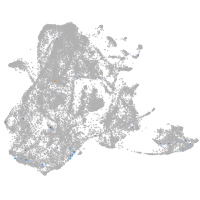 |
periderm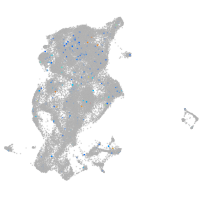 |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| btr22 | 0.099 | tuba1c | -0.031 |
| btg4 | 0.062 | gpm6aa | -0.029 |
| cdca9 | 0.059 | rtn1a | -0.029 |
| buc2l | 0.058 | stmn1b | -0.028 |
| si:dkey-208k4.2 | 0.058 | elavl3 | -0.027 |
| buc | 0.056 | gpm6ab | -0.027 |
| ca15b | 0.053 | nova2 | -0.027 |
| gtf3ab | 0.052 | marcksl1a | -0.027 |
| si:rp71-45k5.2 | 0.052 | atp6v0cb | -0.026 |
| sinup | 0.052 | ckbb | -0.026 |
| zgc:113424 | 0.052 | gng3 | -0.025 |
| dhrs13a.2 | 0.050 | hmgb3a | -0.025 |
| larp6b | 0.050 | rnasekb | -0.024 |
| stard14 | 0.049 | sncb | -0.024 |
| wee2 | 0.049 | vamp2 | -0.023 |
| FO834825.1 | 0.048 | elavl4 | -0.022 |
| mcm3l | 0.048 | fam168a | -0.022 |
| map1lc3c | 0.047 | tmsb | -0.022 |
| bmp15 | 0.046 | atp6v1e1b | -0.021 |
| cyp11a1 | 0.046 | gapdhs | -0.021 |
| elovl7b | 0.046 | hbbe1.3 | -0.021 |
| lsm14b | 0.046 | rtn1b | -0.021 |
| scel | 0.046 | si:dkey-276j7.1 | -0.021 |
| si:ch73-52f15.5 | 0.046 | stx1b | -0.021 |
| spint2 | 0.046 | tuba1a | -0.021 |
| baiap2l1a | 0.045 | tubb5 | -0.021 |
| cldn23a | 0.045 | ywhag2 | -0.021 |
| cth1 | 0.045 | zc4h2 | -0.021 |
| eloal | 0.045 | stxbp1a | -0.021 |
| h2af1al | 0.045 | calm1a | -0.021 |
| pabpn1l | 0.045 | atpv0e2 | -0.020 |
| si:ch1073-340i21.3 | 0.045 | celf2 | -0.020 |
| si:ch211-132b12.8 | 0.045 | cspg5a | -0.020 |
| si:ch211-207n23.2 | 0.045 | fez1 | -0.020 |
| si:ch73-204p21.2 | 0.045 | gnao1a | -0.020 |




