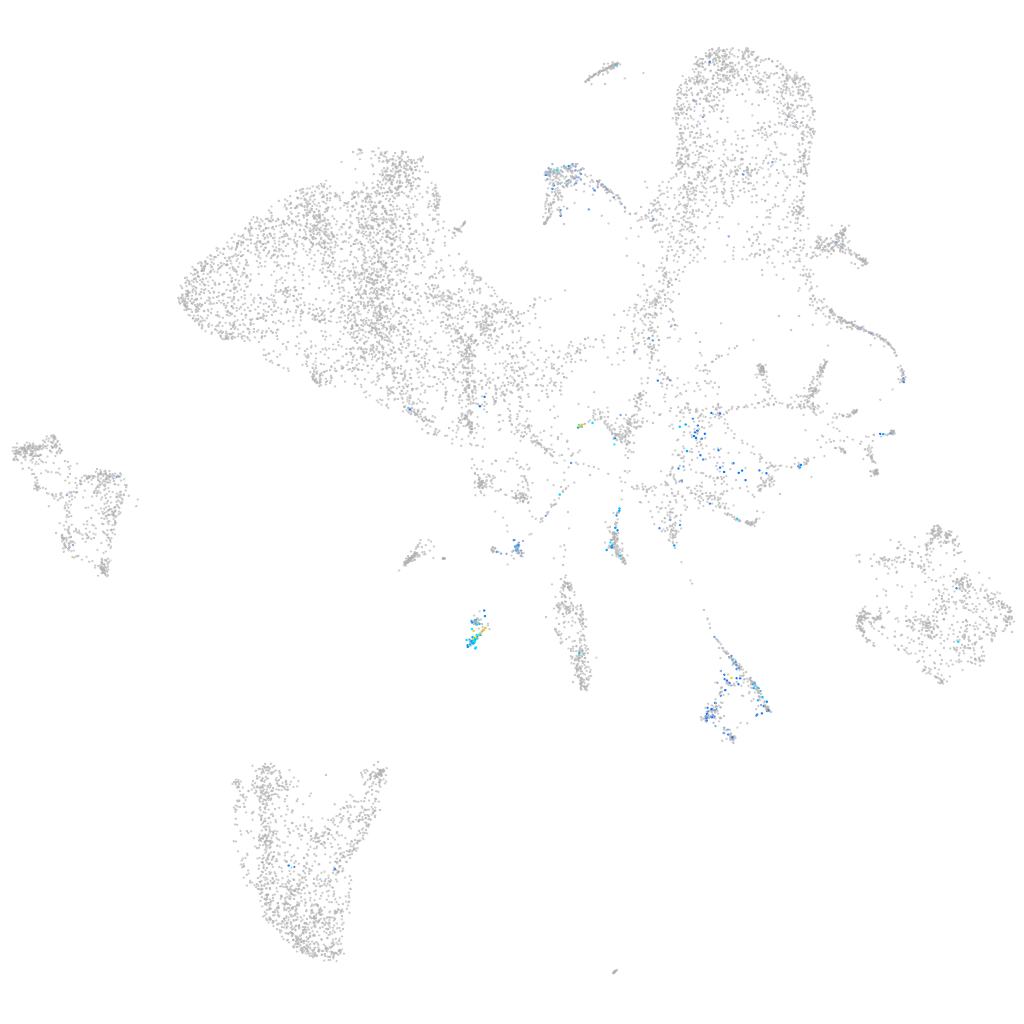
Bruton agammaglobulinemia tyrosine kinase
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:dkey-96g2.1 | 0.461 | gamt | -0.126 |
| zgc:113142 | 0.402 | ahcy | -0.120 |
| cldn2 | 0.355 | fbp1b | -0.120 |
| blnk | 0.350 | aldob | -0.119 |
| ehf | 0.323 | gatm | -0.119 |
| ecm1b | 0.303 | mat1a | -0.114 |
| cav2 | 0.296 | gstt1a | -0.110 |
| cx30.3 | 0.291 | apoa4b.1 | -0.109 |
| zgc:193726 | 0.284 | apoa1b | -0.109 |
| GUCY1A2 | 0.280 | apoc2 | -0.108 |
| pdzk1ip1 | 0.270 | afp4 | -0.106 |
| kctd12.2 | 0.264 | scp2a | -0.106 |
| CR396595.1 | 0.259 | apoc1 | -0.099 |
| marcksl1a | 0.252 | dap | -0.098 |
| entpd8 | 0.249 | mdh1aa | -0.098 |
| cobll1a | 0.249 | agxtb | -0.098 |
| si:ch211-153b23.7 | 0.242 | gpx4a | -0.097 |
| cd9b | 0.236 | apobb.1 | -0.097 |
| cgnb | 0.236 | g6pca.2 | -0.097 |
| gdpd2 | 0.232 | tdo2a | -0.095 |
| cav1 | 0.231 | haao | -0.095 |
| myl9b | 0.230 | si:dkey-16p21.8 | -0.095 |
| rab11fip1a | 0.228 | abat | -0.094 |
| gprc5c | 0.227 | rdh1 | -0.094 |
| rassf10a | 0.226 | gapdh | -0.093 |
| shank3a | 0.224 | msrb2 | -0.092 |
| b3gnt2l | 0.221 | slco1d1 | -0.092 |
| ppp1r9bb | 0.220 | sult2st2 | -0.092 |
| tpm4a | 0.219 | glud1b | -0.092 |
| rassf7b | 0.217 | suclg2 | -0.091 |
| si:dkeyp-82a1.4 | 0.217 | nipsnap3a | -0.089 |
| anxa4 | 0.217 | ttc36 | -0.089 |
| LOC103909050 | 0.216 | serpina1l | -0.089 |
| zgc:198329 | 0.216 | phyhd1 | -0.089 |
| slc4a4b | 0.215 | serpina1 | -0.089 |