basonuclin 2
ZFIN 
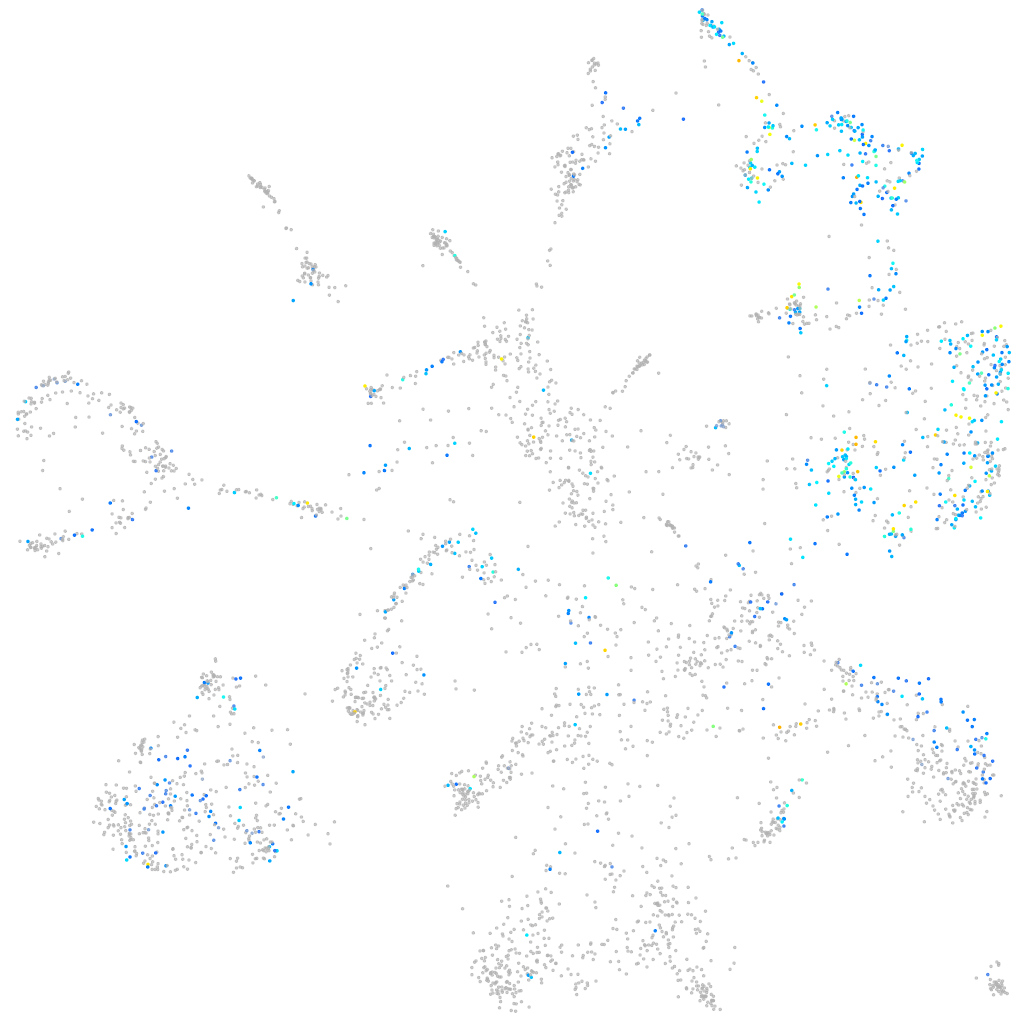
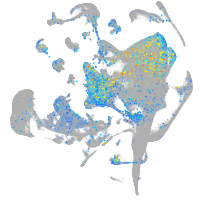
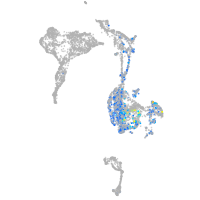
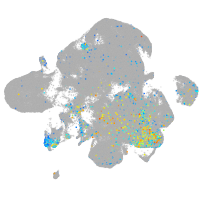
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| podxl | 0.459 | BX088707.3 | -0.270 |
| jam2b | 0.444 | tpm1 | -0.246 |
| tmem88b | 0.401 | acta2 | -0.223 |
| krt94 | 0.392 | myh11a | -0.222 |
| ftr82 | 0.356 | tagln | -0.220 |
| cd151 | 0.348 | myl9a | -0.218 |
| bcam | 0.330 | mylkb | -0.194 |
| aqp1a.1 | 0.327 | myl6 | -0.189 |
| cxadr | 0.327 | lmod1b | -0.180 |
| cavin2b | 0.318 | ptmaa | -0.176 |
| krt8 | 0.317 | inka1a | -0.174 |
| mmel1 | 0.315 | tpm2 | -0.168 |
| gata5 | 0.298 | gapdhs | -0.163 |
| gstm.3 | 0.290 | csrp1b | -0.162 |
| epb41l5 | 0.290 | fhl2a | -0.161 |
| synpo2lb | 0.288 | ak1 | -0.157 |
| bmper | 0.284 | igfbp7 | -0.155 |
| fabp11a | 0.284 | cald1b | -0.155 |
| phactr4a | 0.283 | desmb | -0.152 |
| krt15 | 0.277 | ckbb | -0.151 |
| si:ch211-156j16.1 | 0.276 | cnn1b | -0.151 |
| col12a1a | 0.276 | myocd | -0.149 |
| cav1 | 0.275 | rbpms2a | -0.143 |
| cx43.4 | 0.273 | tmem119b | -0.141 |
| hspb1 | 0.269 | BX323087.1 | -0.138 |
| tgm2b | 0.266 | si:ch211-62a1.3 | -0.134 |
| COLEC10 | 0.266 | atp5l | -0.134 |
| gata6 | 0.265 | ppp1cbl | -0.131 |
| eppk1 | 0.264 | calm2a | -0.131 |
| cavin1b | 0.256 | atp5mc1 | -0.122 |
| tmem98 | 0.252 | cap1 | -0.122 |
| nr0b2a | 0.250 | XLOC-025423 | -0.122 |
| sfrp5 | 0.250 | gpia | -0.122 |
| krt18a.1 | 0.249 | kcne4 | -0.122 |
| tjp2b | 0.245 | cox6b1 | -0.121 |