BMS1 ribosome biogenesis factor
ZFIN 
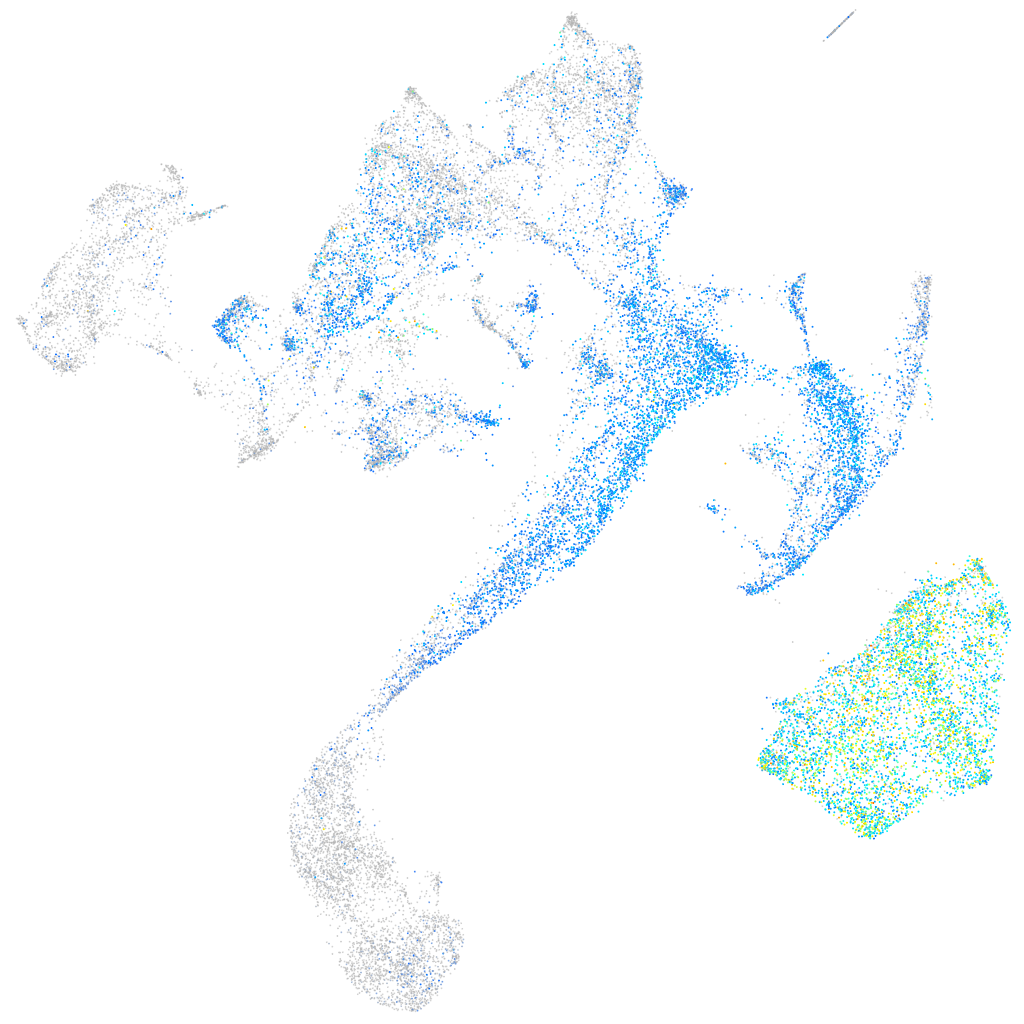
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ncl | 0.689 | rpl37 | -0.588 |
| nop58 | 0.665 | zgc:114188 | -0.543 |
| fbl | 0.660 | rps10 | -0.538 |
| dkc1 | 0.652 | rps17 | -0.515 |
| npm1a | 0.652 | actc1b | -0.450 |
| nop56 | 0.644 | fabp3 | -0.424 |
| anp32e | 0.633 | vdac3 | -0.423 |
| hnrnpa1b | 0.629 | ckma | -0.406 |
| nop2 | 0.623 | ak1 | -0.403 |
| hnrnpub | 0.616 | ckmb | -0.399 |
| ppig | 0.615 | bhmt | -0.399 |
| ilf3b | 0.598 | idh2 | -0.390 |
| ddx18 | 0.597 | tpi1b | -0.389 |
| ebna1bp2 | 0.594 | eno3 | -0.388 |
| gnl3 | 0.594 | tnnc2 | -0.384 |
| nucks1a | 0.594 | atp2a1 | -0.383 |
| hnrnpa1a | 0.592 | eef1da | -0.382 |
| hnrnpabb | 0.592 | atp5meb | -0.380 |
| NC-002333.4 | 0.589 | neb | -0.379 |
| syncrip | 0.589 | mt-nd1 | -0.379 |
| rbm4.3 | 0.585 | aldoab | -0.374 |
| snrnp70 | 0.581 | zgc:158463 | -0.374 |
| seta | 0.581 | pvalb1 | -0.370 |
| top1l | 0.579 | mylpfa | -0.370 |
| acin1a | 0.579 | eif4a1b | -0.367 |
| marcksb | 0.579 | pvalb2 | -0.362 |
| zmp:0000000624 | 0.573 | ttn.2 | -0.361 |
| cdx4 | 0.572 | nme2b.2 | -0.361 |
| srsf1a | 0.572 | ndrg2 | -0.361 |
| srrm1 | 0.570 | eno1a | -0.360 |
| gar1 | 0.566 | ldb3b | -0.360 |
| pes | 0.566 | acta1b | -0.360 |
| rsl1d1 | 0.565 | actn3a | -0.358 |
| hmga1a | 0.565 | si:ch73-367p23.2 | -0.356 |
| hnrnpm | 0.562 | capzb | -0.356 |