"biogenesis of lysosomal organelles complex-1, subunit 1"
ZFIN 
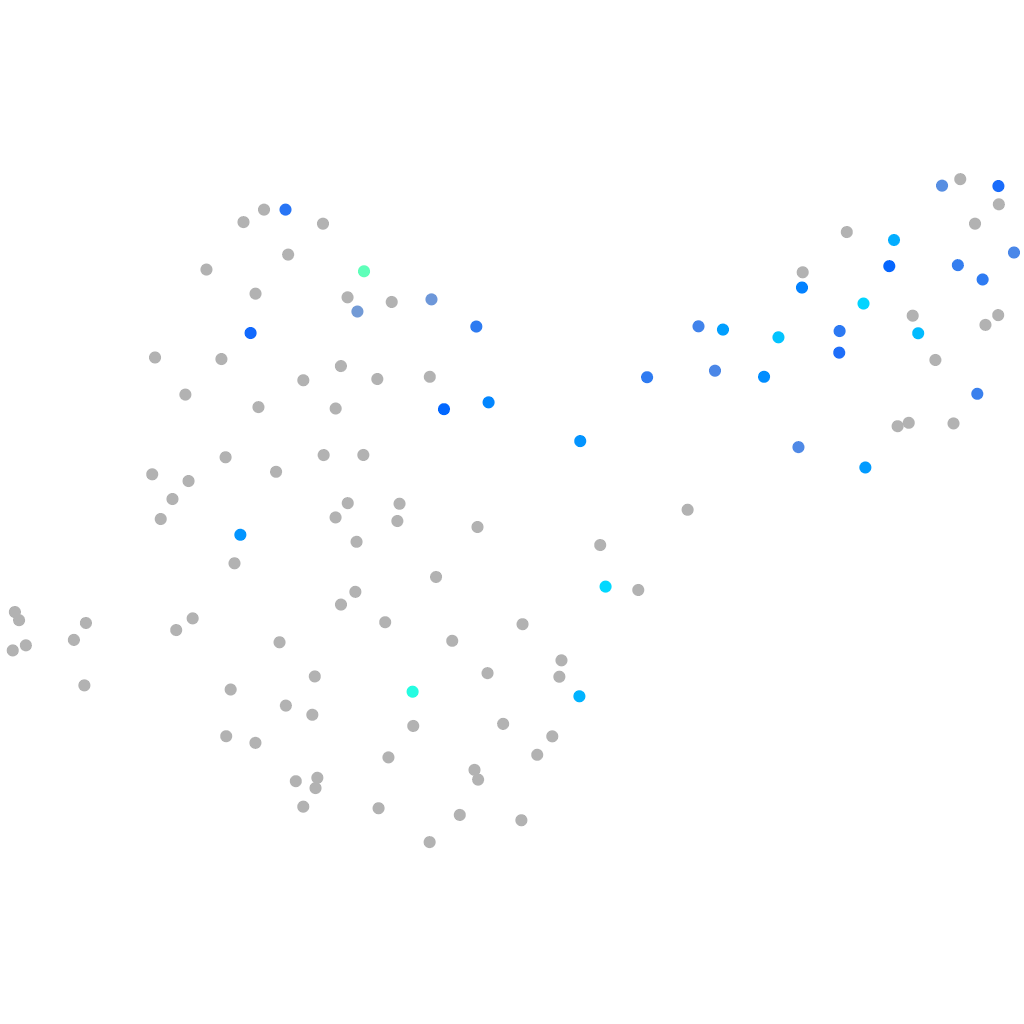
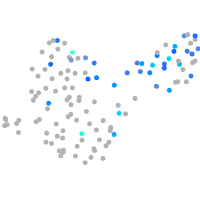
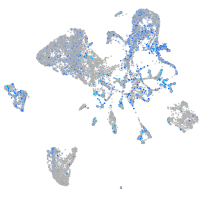
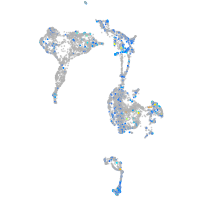
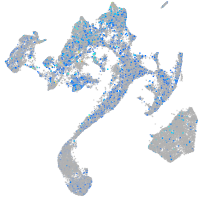
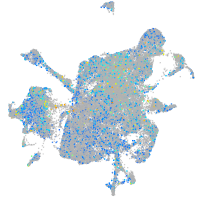
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mbd3b | 0.601 | NC-002333.4 | -0.467 |
| si:ch1073-429i10.3.1 | 0.520 | ncl | -0.458 |
| gapdhs | 0.519 | h1m | -0.409 |
| rtn1a | 0.519 | hnrnpa1b | -0.403 |
| mllt11 | 0.517 | banf1 | -0.403 |
| zgc:114188 | 0.510 | fbl | -0.401 |
| fabp7a | 0.508 | stm | -0.399 |
| fnbp1a | 0.504 | cd2bp2 | -0.395 |
| rpl37 | 0.498 | ppig | -0.389 |
| si:dkey-28b4.7 | 0.498 | hnrnpa1a | -0.383 |
| rps10 | 0.491 | npm1a | -0.383 |
| glod5 | 0.480 | akap12b | -0.381 |
| ggh | 0.478 | anp32e | -0.377 |
| CABZ01038709.1 | 0.474 | ilf3b | -0.377 |
| naa40 | 0.472 | marcksb | -0.373 |
| zbtb16a | 0.471 | ddit4 | -0.373 |
| cnrip1a | 0.470 | hnrnpub | -0.359 |
| tpcn2 | 0.466 | zgc:110425 | -0.358 |
| rps17 | 0.464 | nasp | -0.356 |
| dap1b | 0.464 | nop58 | -0.349 |
| ISCU (1 of many) | 0.464 | hnrnpd | -0.347 |
| pank2 | 0.457 | nucks1a | -0.343 |
| eif1ad | 0.457 | ccna2 | -0.341 |
| dmxl2 | 0.455 | cdk11b | -0.335 |
| slc25a36a | 0.452 | syncrip | -0.335 |
| elmsan1a | 0.449 | lig1 | -0.334 |
| eno1a | 0.448 | hnrnpaba | -0.332 |
| cct2 | 0.448 | ctcf | -0.331 |
| parvb | 0.447 | rps18 | -0.330 |
| emd | 0.447 | rbm7 | -0.329 |
| herc1 | 0.446 | brd3a | -0.327 |
| rps13 | 0.444 | chaf1a | -0.327 |
| tmed4 | 0.444 | snrpa | -0.325 |
| pgam1a | 0.443 | prkrip1 | -0.324 |
| cox6b1 | 0.442 | srrt | -0.324 |