"3-hydroxybutyrate dehydrogenase, type 2"
ZFIN 
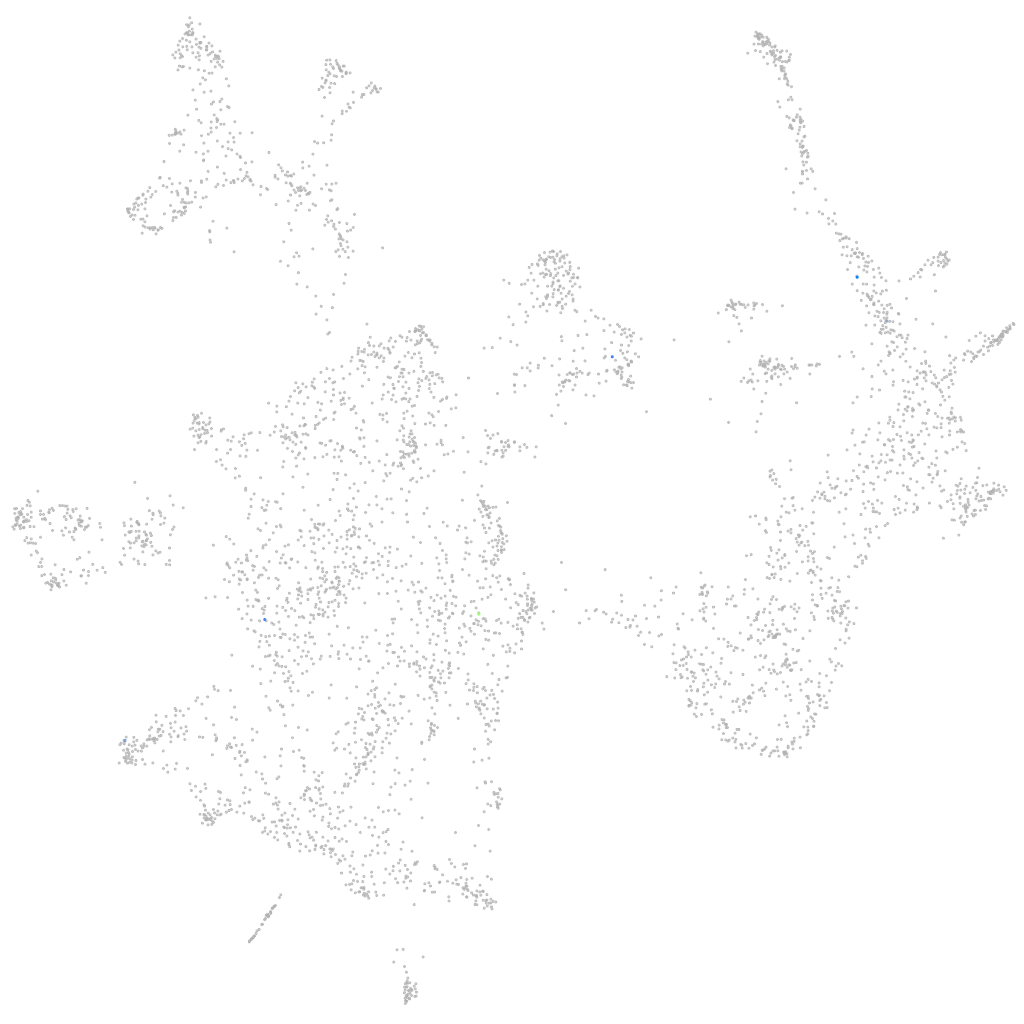
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| zgc:194908 | 0.575 | mt-co1 | -0.044 |
| BX649516.4 | 0.388 | mt-cyb | -0.036 |
| NC-002333.20 | 0.388 | h2afx1 | -0.035 |
| BX255897.1 | 0.332 | fkbp1aa | -0.033 |
| rsad1 | 0.320 | sec61g | -0.032 |
| LOC103909845 | 0.303 | pnrc2 | -0.031 |
| cbsa | 0.277 | slc25a3b | -0.031 |
| fgf1b | 0.264 | oaz1a | -0.030 |
| tigara | 0.264 | lsm6 | -0.028 |
| LOC108180813 | 0.250 | ppib | -0.026 |
| si:ch211-127d4.3 | 0.245 | csnk1a1 | -0.026 |
| LOC101884172 | 0.234 | h3f3a | -0.026 |
| tspan13a | 0.227 | pomp | -0.025 |
| LOC110440045 | 0.183 | syncrip | -0.025 |
| znf1157 | 0.180 | ddx61 | -0.024 |
| DHX35 | 0.172 | jpt1b | -0.024 |
| CR513782.3 | 0.165 | ndufa4l | -0.024 |
| zgc:91944 | 0.165 | cox5aa | -0.024 |
| tnfsf10l4 | 0.160 | ndufs5 | -0.024 |
| slc46a1 | 0.153 | dynll1 | -0.023 |
| malrd1 | 0.151 | eny2 | -0.023 |
| prob1 | 0.147 | jun | -0.023 |
| hsd17b8 | 0.145 | calm2b | -0.023 |
| eef2kmt | 0.139 | epcam | -0.023 |
| LOC101883810 | 0.139 | tmed7 | -0.022 |
| sparcl1 | 0.139 | sub1a | -0.022 |
| CR450848.1 | 0.138 | ctsla | -0.022 |
| si:dkeyp-77c8.2 | 0.138 | si:ch73-46j18.5 | -0.022 |
| lipt1 | 0.137 | mtdha | -0.022 |
| ppic | 0.136 | actb2 | -0.022 |
| hps4 | 0.136 | ewsr1a | -0.022 |
| CR848841.1 | 0.134 | cct6a | -0.022 |
| LOC103908636 | 0.131 | ndufb6 | -0.022 |
| si:cabz01063885.1 | 0.130 | serf2 | -0.022 |
| LOC101885938 | 0.123 | zgc:153713 | -0.022 |