branched chain keto acid dehydrogenase E1 subunit beta
ZFIN 
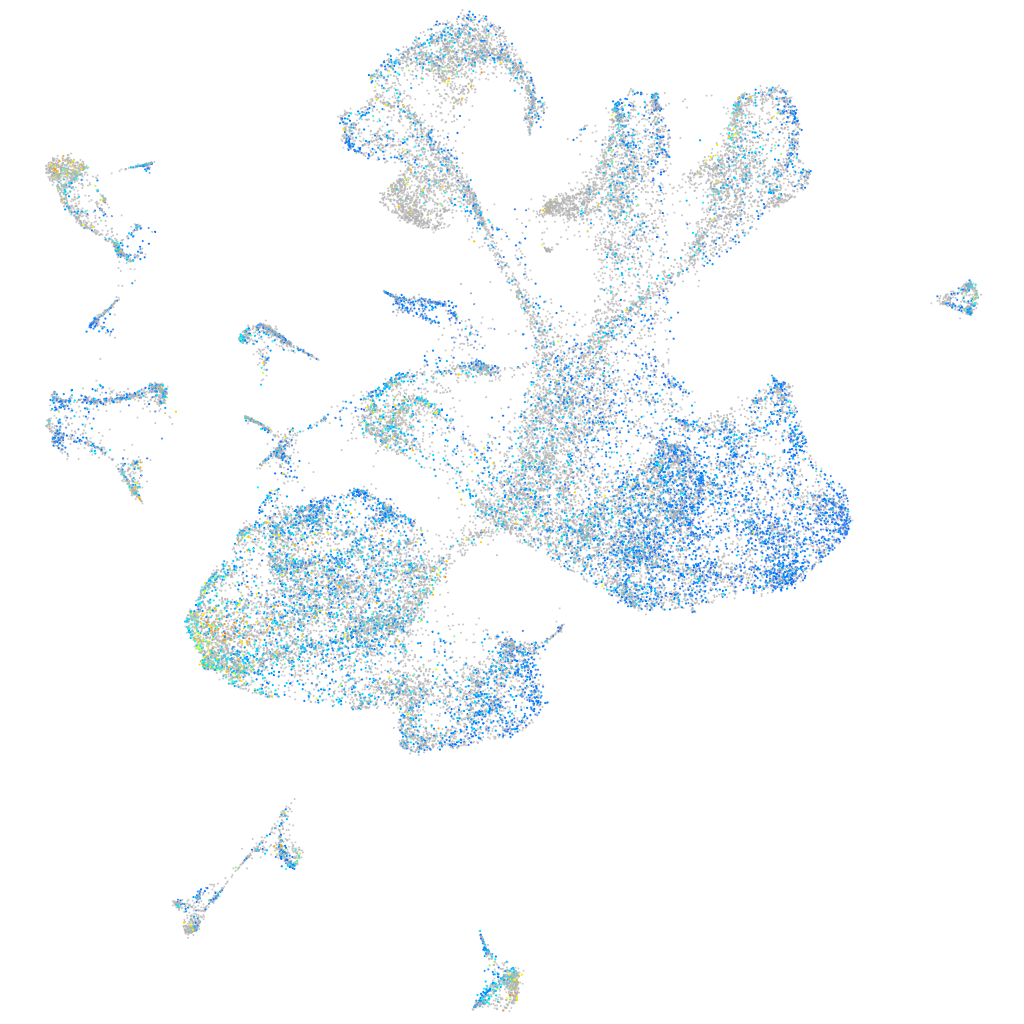
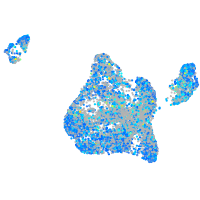
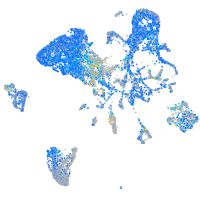
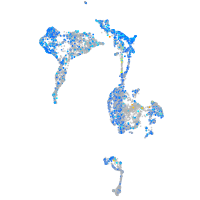
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| atp5mc3b | 0.125 | elavl3 | -0.140 |
| fabp7a | 0.123 | myt1b | -0.117 |
| glula | 0.122 | pik3r3b | -0.107 |
| slc3a2a | 0.121 | tmsb | -0.101 |
| id1 | 0.118 | LOC100537384 | -0.098 |
| atp5f1b | 0.117 | zfhx3 | -0.090 |
| eno1b | 0.116 | rtn1a | -0.088 |
| cx43 | 0.116 | si:ch73-386h18.1 | -0.084 |
| psph | 0.112 | stmn1b | -0.084 |
| si:ch211-66e2.5 | 0.111 | zc4h2 | -0.084 |
| cnbpa | 0.111 | onecut1 | -0.083 |
| atp5pd | 0.111 | myt1a | -0.083 |
| atp5mc1 | 0.111 | ptmab | -0.080 |
| ptgdsb.2 | 0.110 | pou2f2a | -0.079 |
| efhd1 | 0.110 | scrt2 | -0.079 |
| ppap2d | 0.110 | FO082781.1 | -0.079 |
| phgdh | 0.108 | sox11b | -0.078 |
| prdx5 | 0.108 | sox4a | -0.078 |
| GCA | 0.107 | elavl4 | -0.078 |
| uqcrh | 0.106 | nova2 | -0.076 |
| ccng1 | 0.105 | gng2 | -0.074 |
| atp1b4 | 0.105 | nsg2 | -0.072 |
| anxa13 | 0.105 | epb41a | -0.072 |
| atp5pb | 0.105 | baz2ba | -0.071 |
| tuba8l4 | 0.105 | h3f3d | -0.071 |
| psat1 | 0.104 | si:ch211-57n23.4 | -0.071 |
| her6 | 0.104 | ebf3a | -0.071 |
| ptgdsb.1 | 0.104 | zgc:100920 | -0.070 |
| mpc2 | 0.104 | nhlh2 | -0.068 |
| cd63 | 0.103 | onecut2 | -0.068 |
| slc4a4a | 0.103 | si:ch211-222l21.1 | -0.068 |
| si:ch211-286b5.5 | 0.103 | marcksl1b | -0.067 |
| ddt | 0.102 | rbfox2 | -0.067 |
| mfge8a | 0.102 | hmgb3a | -0.066 |
| pa2g4b | 0.101 | golga7ba | -0.066 |