"BCAR1 scaffold protein, Cas family member"
ZFIN 
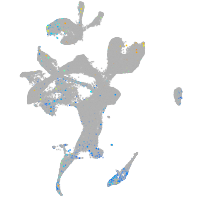
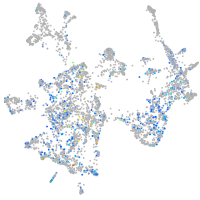
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| marcksl1b | 0.063 | rbp4 | -0.043 |
| NC-002333.4 | 0.054 | fgfbp2b | -0.041 |
| CABZ01075068.1 | 0.054 | col2a1a | -0.041 |
| tmem88b | 0.054 | apoa2 | -0.041 |
| mdka | 0.052 | col9a1a | -0.038 |
| scarb2a | 0.052 | cnmd | -0.037 |
| cdh2 | 0.051 | col9a3 | -0.036 |
| arvcfb | 0.051 | eef1da | -0.036 |
| phospho1 | 0.050 | col9a2 | -0.034 |
| foxd3 | 0.050 | VIT | -0.034 |
| fstl1a | 0.050 | pvalb2 | -0.033 |
| qkia | 0.049 | CABZ01092746.1 | -0.033 |
| ifitm5 | 0.049 | hpdb | -0.033 |
| si:dkey-56m19.5 | 0.048 | hgd | -0.032 |
| rac1a | 0.047 | zgc:153704 | -0.032 |
| dlc1 | 0.047 | pvalb1 | -0.031 |
| tmem119a | 0.046 | apoa1b | -0.030 |
| erbb3b | 0.046 | matn1 | -0.028 |
| cdh7b | 0.046 | sox9a | -0.028 |
| actb1 | 0.046 | ifitm1 | -0.027 |
| tpm4a | 0.046 | otos | -0.027 |
| col5a2b | 0.045 | loxl1 | -0.027 |
| akap12b | 0.045 | aldob | -0.027 |
| slc8a4b | 0.044 | prss1 | -0.027 |
| mcamb | 0.044 | barx1 | -0.026 |
| mxd4 | 0.044 | mgst1.2 | -0.026 |
| gata5 | 0.044 | bhmt | -0.026 |
| col7a1l | 0.044 | slc3a2b | -0.026 |
| pleca | 0.044 | eif4ebp3 | -0.025 |
| marcksl1a | 0.043 | rsl24d1 | -0.025 |
| tmsb4x | 0.043 | cdkn1a | -0.025 |
| nr2f5 | 0.043 | prss59.2 | -0.025 |
| lin28a | 0.043 | nme2b.2 | -0.025 |
| lsp1 | 0.043 | snorc | -0.025 |
| CABZ01072614.1 | 0.042 | mia | -0.025 |