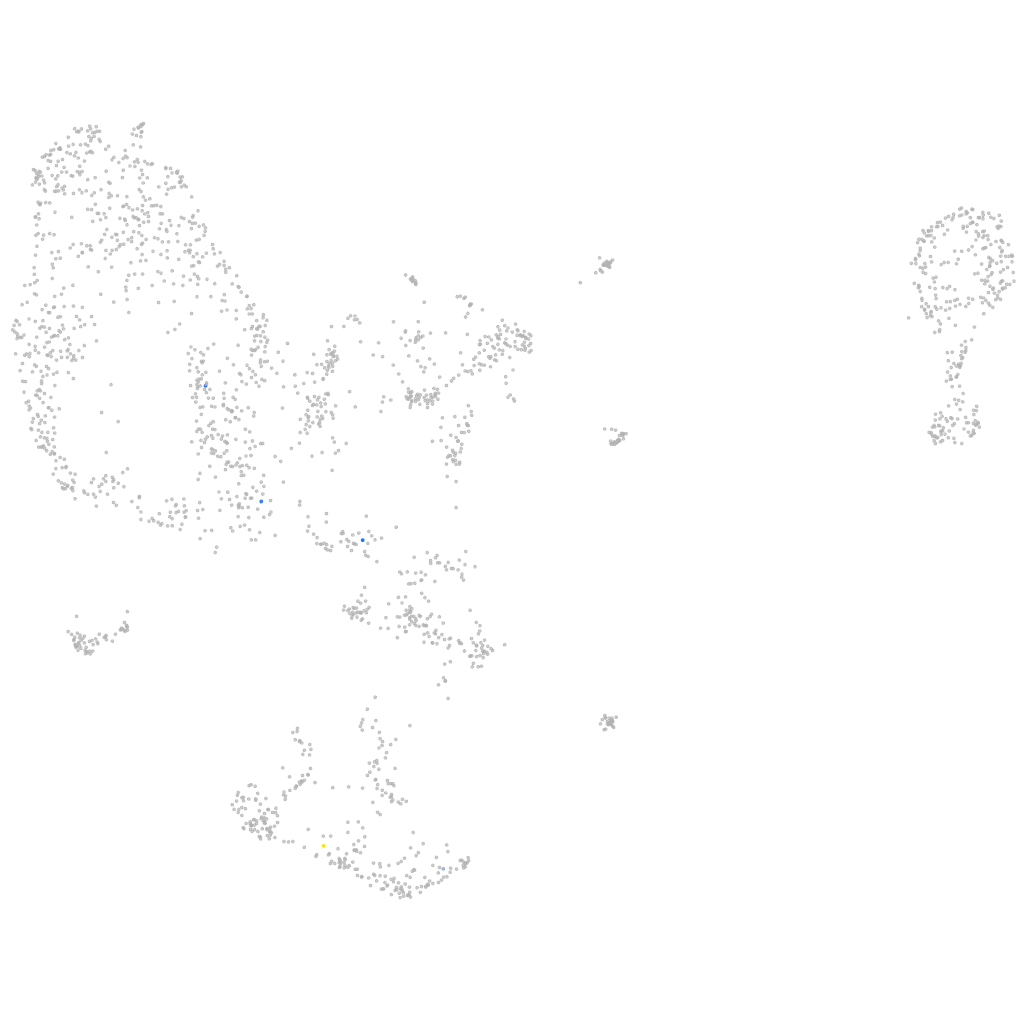
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| uckl1a | 0.821 | btf3 | -0.069 |
| rlbp1a | 0.717 | eif3f | -0.047 |
| pcdh1gc5 | 0.705 | sec61g | -0.045 |
| insl5a | 0.580 | rpl18a | -0.045 |
| csf1b | 0.577 | fthl27 | -0.043 |
| LO018608.1 | 0.575 | zgc:193541 | -0.042 |
| pou4f3 | 0.560 | rsl24d1 | -0.036 |
| abi3a | 0.555 | bsg | -0.034 |
| CABZ01055522.1 | 0.524 | pnrc2 | -0.032 |
| thbs2b | 0.486 | ubb | -0.032 |
| arr3a | 0.462 | mt-nd5 | -0.032 |
| tmx3a | 0.457 | sdhc | -0.032 |
| crabp1a | 0.428 | eif2s3 | -0.031 |
| LOC101886429 | 0.422 | zgc:111986 | -0.031 |
| opn1mw1 | 0.413 | ctsla | -0.030 |
| nrgna | 0.389 | myl12.1 | -0.030 |
| arr3b | 0.384 | gabarapb | -0.030 |
| vstm4a | 0.383 | eif1b | -0.030 |
| cdh26.1 | 0.380 | rpl22l1 | -0.030 |
| rfx7a | 0.379 | ndufs6 | -0.030 |
| prom1b | 0.376 | atox1 | -0.030 |
| si:dkeyp-73d8.9 | 0.366 | rbm8a | -0.030 |
| c2cd4a | 0.355 | sem1 | -0.029 |
| zgc:171482 | 0.348 | atp1a1a.1 | -0.029 |
| ckmt2a | 0.346 | khdrbs1a | -0.029 |
| fhdc4 | 0.342 | sat1a.2 | -0.029 |
| caskin1 | 0.335 | ndufb7 | -0.029 |
| gdf6a | 0.329 | si:dkey-56m19.5 | -0.028 |
| tnni1c | 0.312 | eif5 | -0.028 |
| atg4db | 0.299 | ncl | -0.028 |
| LOC110439513 | 0.299 | mibp2 | -0.028 |
| si:ch211-212k18.9 | 0.287 | ccdc25 | -0.028 |
| cthrc1a | 0.279 | psmd14 | -0.028 |
| smarcd3a | 0.277 | hnrnpub | -0.028 |
| mapk8ip3 | 0.267 | cldnb | -0.028 |