Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis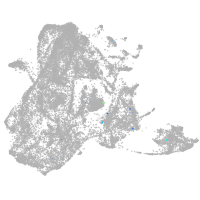 |
periderm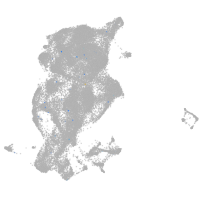 |
fin |
ionocytes / mucous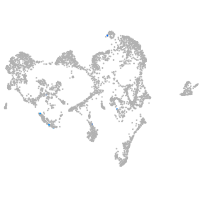 |
pigment cells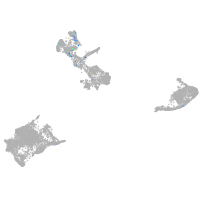 |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CT737162.3 | 0.054 | aldob | -0.013 |
| si:ch211-38m6.6 | 0.052 | hspb1 | -0.012 |
| hsf5 | 0.051 | lig1 | -0.012 |
| si:ch211-256m1.8 | 0.051 | polr3gla | -0.012 |
| si:ch73-89b15.3 | 0.048 | akap12b | -0.011 |
| unm-sa821 | 0.048 | dkc1 | -0.011 |
| gpnmb | 0.047 | fkbp7 | -0.011 |
| tmem179b | 0.046 | pou5f3 | -0.011 |
| apoda.1 | 0.045 | tbx16 | -0.011 |
| akap12a | 0.044 | zic2b | -0.011 |
| CR318624.2 | 0.044 | actc1b | -0.010 |
| fhl2b | 0.044 | apoc1 | -0.010 |
| lypc | 0.044 | banf1 | -0.010 |
| defbl1 | 0.042 | col1a2 | -0.010 |
| zgc:110789 | 0.039 | cyt1 | -0.010 |
| ltk | 0.038 | cyt1l | -0.010 |
| ak9 | 0.038 | fbxo5 | -0.010 |
| cfap52 | 0.037 | id1 | -0.010 |
| si:dkey-197i20.6 | 0.037 | msna | -0.010 |
| ccdc151 | 0.036 | nnr | -0.010 |
| spag6 | 0.036 | pvalb1 | -0.010 |
| tekt1 | 0.036 | pvalb2 | -0.010 |
| ednrba | 0.035 | si:ch1073-80i24.3 | -0.010 |
| gmnc | 0.035 | si:ch211-152c2.3 | -0.010 |
| mycbpap | 0.035 | si:dkey-66i24.9 | -0.010 |
| pnp4a | 0.035 | ved | -0.010 |
| si:dkey-30c15.17 | 0.035 | vox | -0.010 |
| casc1 | 0.034 | add3b | -0.009 |
| ccdc169 | 0.034 | asb11 | -0.009 |
| CFAP77 | 0.034 | avd | -0.009 |
| slc25a38a | 0.034 | ccna2 | -0.009 |
| smkr1 | 0.034 | cdca7a | -0.009 |
| cfap126 | 0.033 | chaf1a | -0.009 |
| CR847897.1 | 0.033 | ckap4 | -0.009 |
| dnaaf1 | 0.033 | col1a1a | -0.009 |




