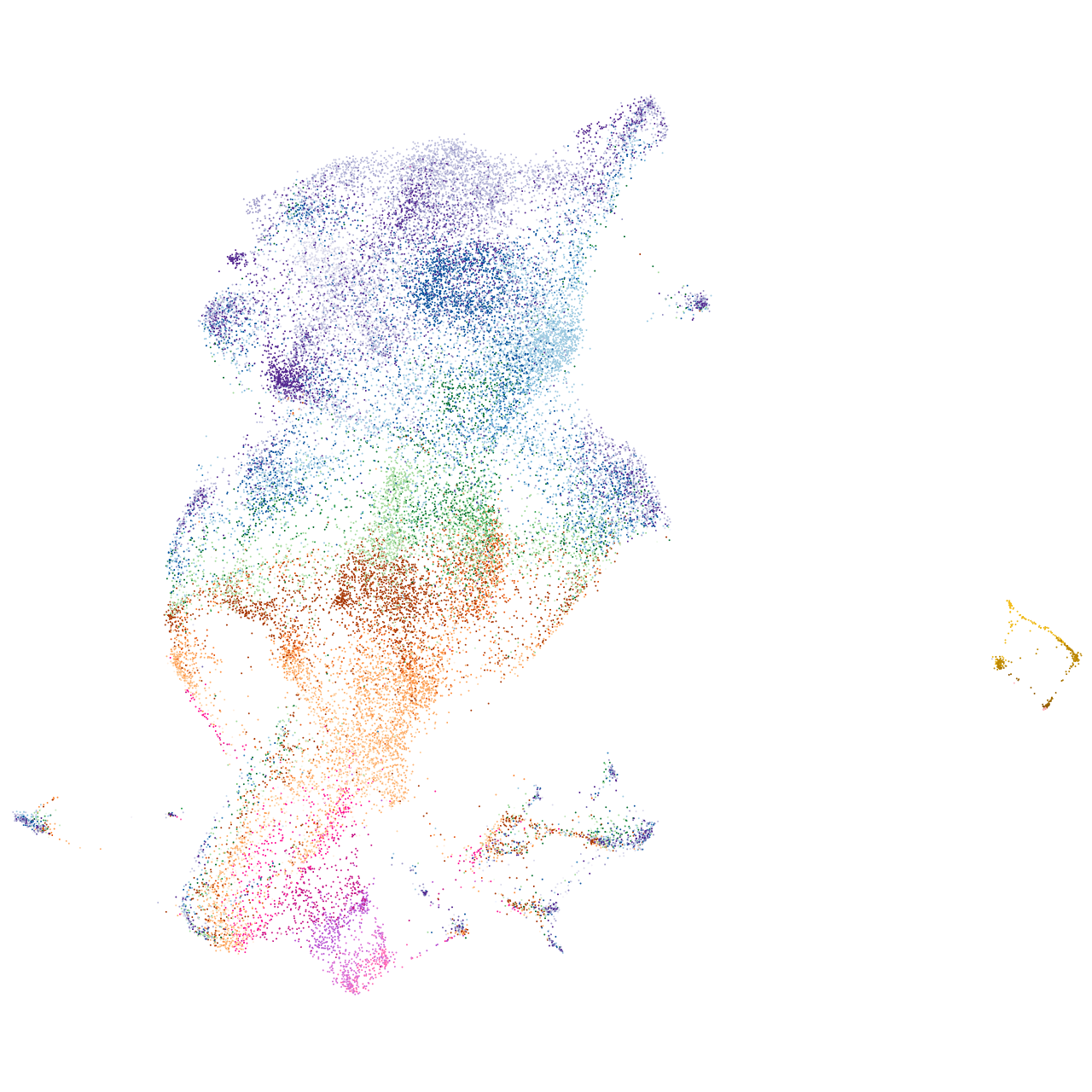Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory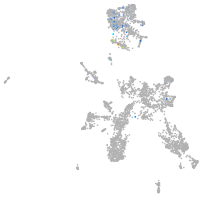 |
otic / lateral line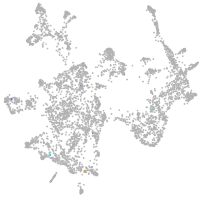 |
epidermis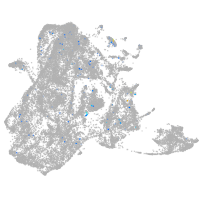 |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| BX928743.1 | 0.316 | krt4 | -0.017 |
| si:dkey-85k15.4 | 0.263 | zgc:193505 | -0.015 |
| CR786571.3 | 0.243 | agr1 | -0.015 |
| zgc:109913 | 0.230 | anxa1c | -0.015 |
| hepacam2 | 0.227 | cyt1l | -0.015 |
| cngb3.2 | 0.226 | gsnb | -0.014 |
| grp | 0.224 | eif3ja | -0.014 |
| ccdc39 | 0.213 | zgc:175088 | -0.014 |
| ano8a | 0.212 | krt92 | -0.014 |
| KCNJ6 | 0.204 | tmem189 | -0.013 |
| LOC101887111 | 0.203 | tmed10 | -0.013 |
| st3gal1l | 0.198 | cfl1l | -0.013 |
| CABZ01073083.1 | 0.197 | fbl | -0.012 |
| zgc:112183 | 0.188 | krt5 | -0.012 |
| si:ch211-256e16.6 | 0.188 | commd2 | -0.012 |
| cldn8 | 0.187 | ssr4 | -0.012 |
| CR759836.1 | 0.185 | prkar2aa | -0.012 |
| CU137681.8 | 0.184 | sap18 | -0.012 |
| igf1 | 0.183 | ddost | -0.012 |
| tyrobp | 0.182 | arpc1a | -0.012 |
| si:ch211-225k7.5 | 0.179 | zgc:153284 | -0.011 |
| LOC798089 | 0.172 | tmem258 | -0.011 |
| tmem163b | 0.169 | rgcc | -0.011 |
| otofa | 0.167 | prickle1b | -0.011 |
| cldnh | 0.165 | kdelr2a | -0.011 |
| cers3b | 0.160 | atp5pb | -0.011 |
| il1rl1 | 0.155 | slc25a55a | -0.011 |
| si:ch211-266g18.6 | 0.142 | tomm5 | -0.011 |
| sytl2a | 0.139 | naa15a | -0.011 |
| smap2 | 0.138 | abracl | -0.010 |
| chga | 0.137 | ube2na | -0.010 |
| prr15la | 0.136 | nup93 | -0.010 |
| mmp17a | 0.136 | cxcl14 | -0.010 |
| zgc:101744 | 0.135 | wu:fb18f06 | -0.010 |
| si:ch211-278j3.3 | 0.134 | ccdc25 | -0.010 |