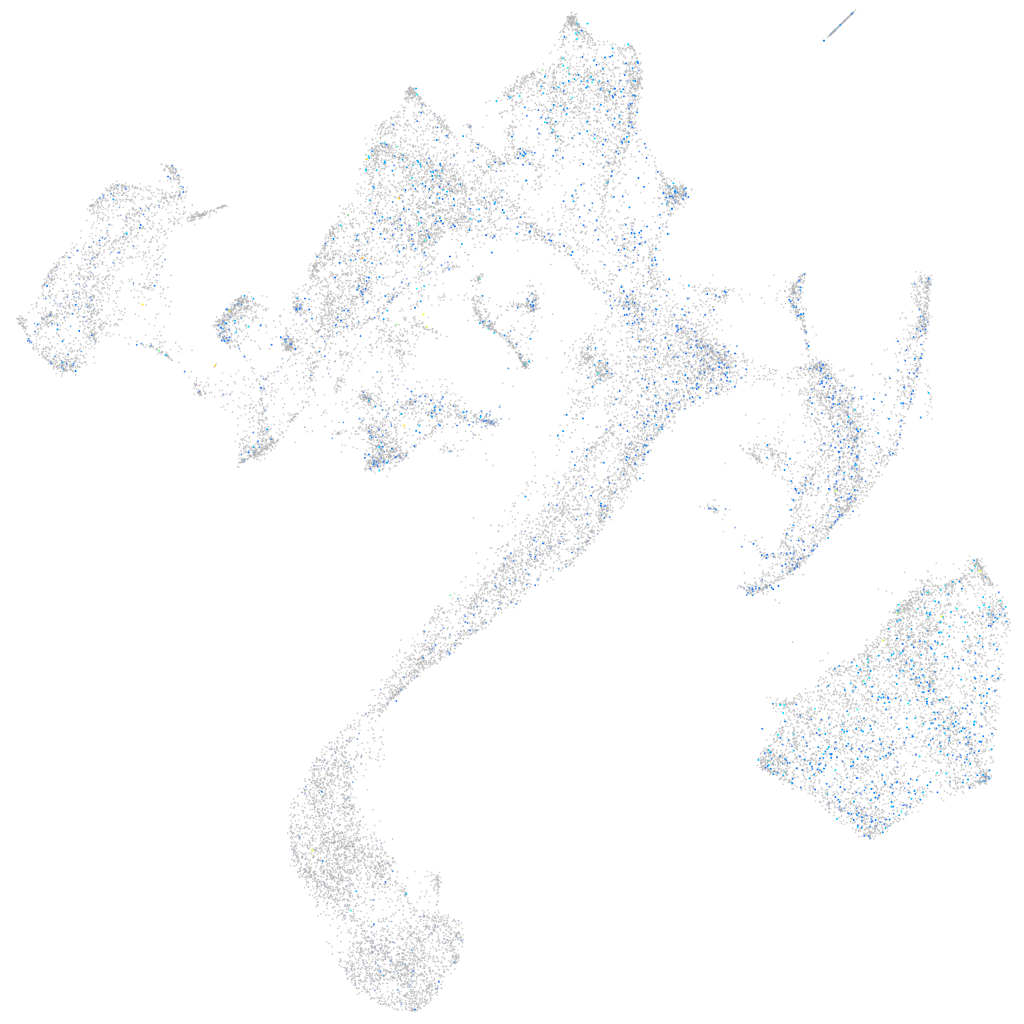
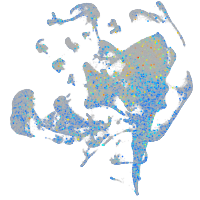
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch211-222l21.1 | 0.117 | actc1b | -0.103 |
| cirbpa | 0.113 | atp2a1 | -0.090 |
| hnrnpa1b | 0.110 | ckmb | -0.090 |
| ptmab | 0.109 | ak1 | -0.089 |
| anp32b | 0.109 | tpma | -0.089 |
| hmga1a | 0.109 | tnnc2 | -0.088 |
| smc1al | 0.108 | ckma | -0.088 |
| hmgb2a | 0.108 | tmem38a | -0.088 |
| chaf1a | 0.108 | mylpfa | -0.088 |
| seta | 0.108 | acta1b | -0.087 |
| si:ch211-288g17.3 | 0.107 | ttn.2 | -0.087 |
| marcksb | 0.107 | aldoab | -0.087 |
| hnrnpa1a | 0.106 | gamt | -0.086 |
| nucks1a | 0.106 | mylz3 | -0.085 |
| si:ch73-1a9.3 | 0.104 | myl1 | -0.085 |
| top1l | 0.103 | mybphb | -0.083 |
| acin1a | 0.103 | neb | -0.083 |
| hnrnpabb | 0.103 | si:ch73-367p23.2 | -0.081 |
| banf1 | 0.102 | tnnt3a | -0.081 |
| ppm1g | 0.102 | pabpc4 | -0.081 |
| hnrnpaba | 0.102 | ttn.1 | -0.081 |
| syncrip | 0.102 | cav3 | -0.080 |
| cbx1a | 0.102 | srl | -0.079 |
| hmgb2b | 0.102 | ldb3a | -0.079 |
| smarca4a | 0.101 | pvalb2 | -0.078 |
| si:ch73-281n10.2 | 0.101 | CABZ01078594.1 | -0.078 |
| khdrbs1a | 0.101 | actn3b | -0.078 |
| hnrnpa0b | 0.101 | txlnbb | -0.078 |
| cx43.4 | 0.101 | actn3a | -0.078 |
| anp32a | 0.101 | hhatla | -0.077 |
| rbbp4 | 0.100 | ldb3b | -0.077 |
| lig1 | 0.100 | ryr1b | -0.077 |
| ctnnb1 | 0.100 | eno1a | -0.077 |
| taf15 | 0.099 | pvalb1 | -0.077 |
| hmgn7 | 0.099 | gapdh | -0.076 |