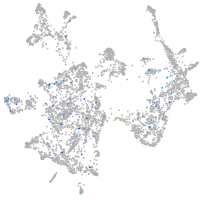
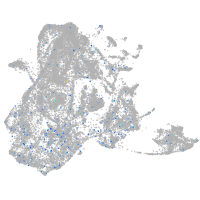
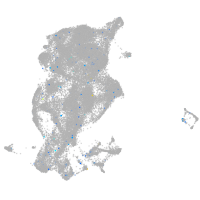
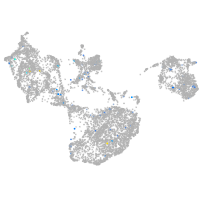
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| BX927216.1 | 0.212 | gamt | -0.068 |
| PRMT8 | 0.183 | bhmt | -0.060 |
| LOC108190923 | 0.168 | apoa2 | -0.058 |
| CR318673.1 | 0.159 | apoa1b | -0.057 |
| LOC100331291 | 0.156 | gatm | -0.056 |
| sh2d4ba | 0.154 | pnp4b | -0.056 |
| BX088603.1 | 0.144 | agxtb | -0.055 |
| CABZ01068208.1 | 0.139 | rbp4 | -0.054 |
| LOC100332699 | 0.139 | grhprb | -0.053 |
| CR339041.1 | 0.135 | gapdh | -0.052 |
| lhb | 0.134 | ahcy | -0.052 |
| TMC1 | 0.133 | zgc:123103 | -0.051 |
| si:dkey-171c9.3 | 0.131 | mat1a | -0.051 |
| CABZ01081780.1 | 0.130 | apom | -0.051 |
| LOC103910179 | 0.127 | fetub | -0.051 |
| LOC108180112 | 0.124 | zgc:92744 | -0.050 |
| LOC101885413 | 0.123 | gpx4a | -0.050 |
| LOC101885611 | 0.121 | serpina1 | -0.050 |
| CR931782.2 | 0.120 | fabp10a | -0.049 |
| coch | 0.120 | rbp2b | -0.049 |
| cacna1eb | 0.118 | serpina1l | -0.049 |
| znf1055 | 0.116 | ces2 | -0.049 |
| LOC101882509 | 0.113 | hao1 | -0.048 |
| AL928920.3 | 0.113 | tfa | -0.048 |
| NABP2 | 0.112 | apoa4b.1 | -0.048 |
| LOC100005923 | 0.110 | fgg | -0.047 |
| dennd2db | 0.109 | LOC110437731 | -0.047 |
| LOC100537992 | 0.108 | c9 | -0.047 |
| LOC110439611 | 0.107 | gc | -0.047 |
| rbm46 | 0.106 | ambp | -0.047 |
| pimr201 | 0.105 | ttr | -0.046 |
| XLOC-016099 | 0.104 | afp4 | -0.046 |
| dusp3a | 0.104 | kng1 | -0.046 |
| LOC101884985 | 0.103 | fgb | -0.046 |
| AL929396.1 | 0.103 | f2 | -0.046 |