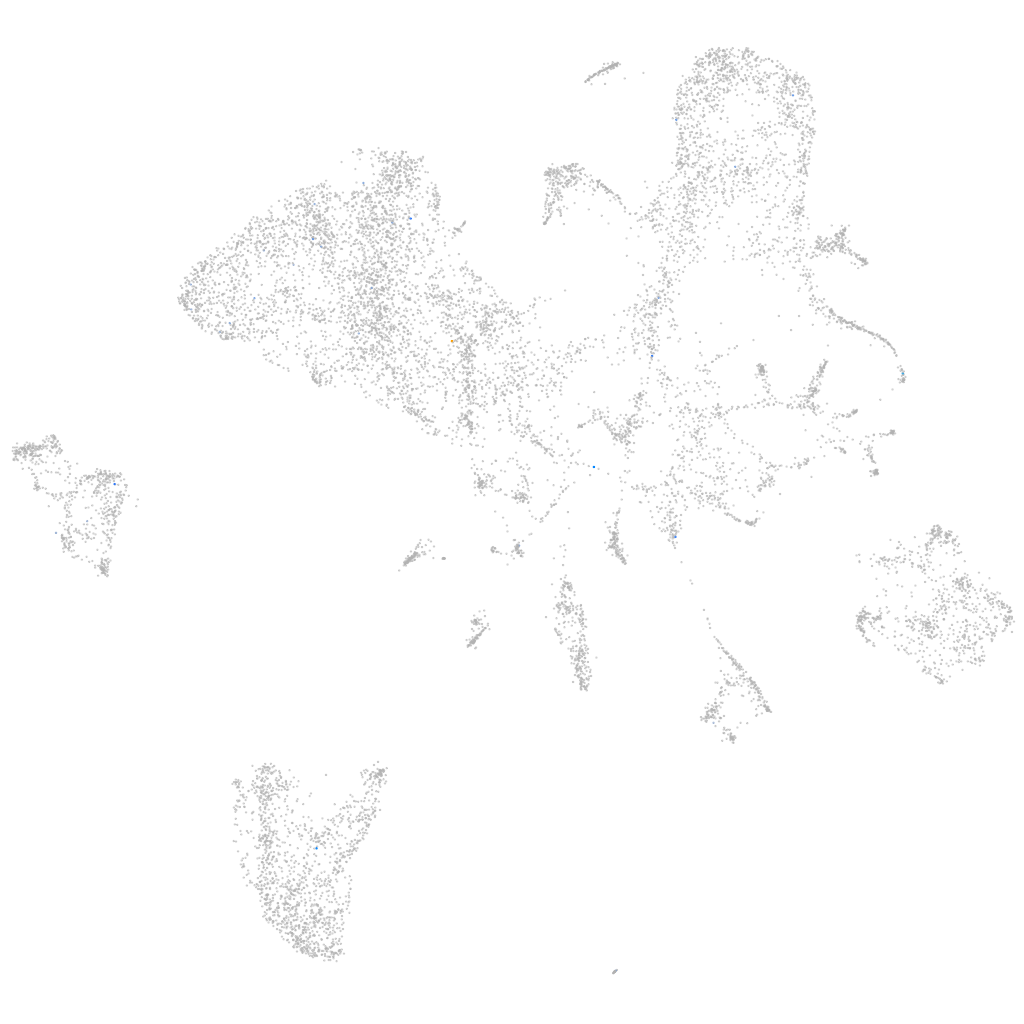
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gabra6a | 0.609 | tpt1 | -0.026 |
| irx4a | 0.319 | sec61g | -0.025 |
| LOC110438090 | 0.273 | rpl36 | -0.025 |
| ropn1l | 0.265 | rpl34 | -0.023 |
| SCCPDH (1 of many) | 0.215 | rps29 | -0.023 |
| XLOC-013086 | 0.190 | cox8a | -0.018 |
| acot7 | 0.169 | oaz1a | -0.017 |
| dock3 | 0.165 | NC-002333.4 | -0.017 |
| BX927394.3 | 0.165 | krtcap2 | -0.017 |
| CU929426.1 | 0.161 | slc25a3b | -0.017 |
| osbpl7 | 0.152 | rpl23a | -0.017 |
| il17rc | 0.148 | mt-cyb | -0.016 |
| fabp11b | 0.143 | erh | -0.016 |
| smyd1a | 0.129 | rpl15 | -0.016 |
| twist3 | 0.123 | arf2b | -0.016 |
| si:ch211-125o16.4 | 0.118 | COX3 | -0.016 |
| dnal4a | 0.101 | rps27.2 | -0.016 |
| cry-dash | 0.100 | atp5mc3b | -0.015 |
| nemp2 | 0.100 | pabpc1a | -0.015 |
| znf1103 | 0.097 | rtf1 | -0.015 |
| ugt5g1 | 0.096 | nfyba | -0.015 |
| EARS2 | 0.095 | cptp | -0.015 |
| rusc2 | 0.093 | cox7b | -0.015 |
| si:dkey-269o24.1 | 0.089 | rnaseka | -0.015 |
| proza | 0.089 | hmgn6 | -0.015 |
| ptpa | 0.088 | chtopa | -0.015 |
| gaa3-as | 0.087 | spcs2 | -0.015 |
| si:ch1073-357b18.4 | 0.086 | tmed2 | -0.015 |
| BX294181.1 | 0.086 | rpl3 | -0.015 |
| sult1st4 | 0.086 | nansa | -0.014 |
| LOC101887034 | 0.086 | rplp2 | -0.014 |
| BX119926.1 | 0.085 | ppib | -0.014 |
| arsj | 0.084 | cirbpb | -0.014 |
| CU207281.1 | 0.084 | slc39a7 | -0.014 |
| bcl11aa | 0.083 | clta | -0.014 |