Other cell groups


all cells |
blastomeres |
PGCs |
endoderm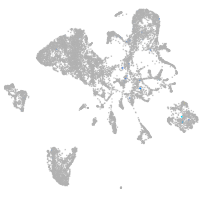 |
axial mesoderm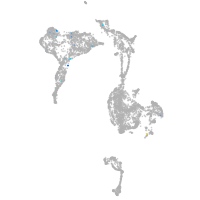 |
muscle 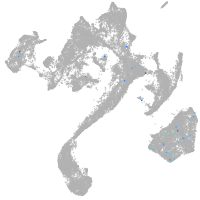 |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis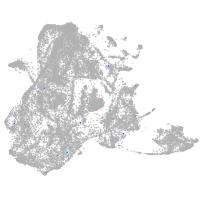 |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| AL954672.1 | 0.112 | rpl37 | -0.040 |
| LOC110439537 | 0.065 | rps10 | -0.039 |
| si:ch211-152c2.3 | 0.053 | ptmaa | -0.039 |
| stm | 0.053 | zgc:114188 | -0.036 |
| hspb1 | 0.050 | h3f3a | -0.033 |
| BX005058.1 | 0.049 | tmsb4x | -0.032 |
| BX296541.1 | 0.049 | hnrnpa0l | -0.031 |
| entpd2b | 0.047 | tuba1c | -0.028 |
| LOC101886161 | 0.047 | fabp3 | -0.027 |
| apoeb | 0.046 | ppiab | -0.026 |
| bms1 | 0.046 | marcksl1a | -0.026 |
| si:dkey-108k21.14 | 0.046 | CR383676.1 | -0.026 |
| si:dkeyp-69c1.6 | 0.045 | rps17 | -0.026 |
| NC-002333.4 | 0.045 | rtn1a | -0.025 |
| crabp2b | 0.045 | stmn1b | -0.025 |
| COX7A2 | 0.044 | gpm6aa | -0.023 |
| fbl | 0.043 | elavl3 | -0.023 |
| nop2 | 0.043 | gpm6ab | -0.022 |
| gar1 | 0.043 | si:ch1073-429i10.3.1 | -0.022 |
| CR376741.4 | 0.043 | mt-nd1 | -0.022 |
| dkc1 | 0.043 | h3f3c | -0.021 |
| nop58 | 0.042 | cct2 | -0.021 |
| galnt7 | 0.042 | mt-atp6 | -0.021 |
| zgc:110425 | 0.042 | pvalb1 | -0.021 |
| si:dkey-66i24.9 | 0.042 | rnasekb | -0.021 |
| kri1 | 0.042 | calm1a | -0.021 |
| pou5f3 | 0.041 | ckbb | -0.021 |
| rrs1 | 0.041 | nme2b.1 | -0.020 |
| dnmt3bb.2 | 0.041 | tmsb | -0.020 |
| vrtn | 0.041 | zgc:158463 | -0.020 |
| alcamb | 0.040 | celf2 | -0.019 |
| npm1a | 0.040 | COX3 | -0.019 |
| akap12b | 0.040 | elavl4 | -0.019 |
| CR847793.1 | 0.040 | sncb | -0.018 |
| CU013526.1 | 0.040 | tubb5 | -0.018 |




