Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line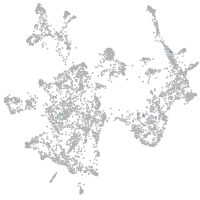 |
epidermis |
periderm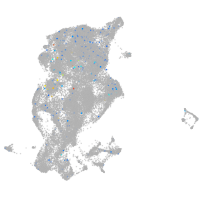 |
fin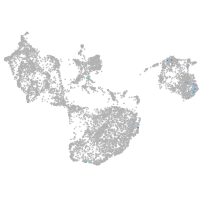 |
ionocytes / mucous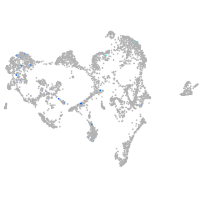 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| dhrs13a.2 | 0.058 | marcksl1a | -0.032 |
| si:ch211-207n23.2 | 0.056 | tuba1c | -0.031 |
| si:ch73-52f15.5 | 0.056 | nova2 | -0.028 |
| si:dkey-247k7.2 | 0.056 | gpm6aa | -0.027 |
| si:dkeyp-1h4.6 | 0.055 | hmgb3a | -0.026 |
| aldob | 0.054 | rtn1a | -0.026 |
| abcb5 | 0.053 | gpm6ab | -0.025 |
| spint2 | 0.053 | hbbe1.3 | -0.025 |
| zgc:111983 | 0.053 | mdkb | -0.025 |
| mid1ip1a | 0.052 | atp6v0cb | -0.024 |
| eevs | 0.051 | ckbb | -0.024 |
| scel | 0.051 | elavl3 | -0.024 |
| pou5f3 | 0.050 | fabp3 | -0.024 |
| rhcgl1 | 0.050 | fam168a | -0.024 |
| stx11b.1 | 0.050 | hbae3 | -0.024 |
| apodb | 0.049 | stmn1b | -0.024 |
| rhcga | 0.049 | hbae1.1 | -0.023 |
| si:dkeyp-51b9.3 | 0.049 | tuba1a | -0.023 |
| stard14 | 0.049 | gapdhs | -0.022 |
| cldne | 0.048 | gng3 | -0.022 |
| krt17 | 0.048 | hmgb1b | -0.022 |
| soul2 | 0.048 | rnasekb | -0.022 |
| tcnbb | 0.048 | wu:fb55g09 | -0.022 |
| tmem176l.4 | 0.048 | cspg5a | -0.021 |
| selenow2b | 0.047 | hbbe1.1 | -0.021 |
| si:ch1073-80i24.3 | 0.047 | sncb | -0.021 |
| si:ch211-79k12.1 | 0.047 | tmeff1b | -0.021 |
| tlcd1 | 0.047 | celf2 | -0.020 |
| XLOC-011066 | 0.047 | h2afy2 | -0.020 |
| cd9b | 0.046 | tubb5 | -0.020 |
| fuca1.1 | 0.046 | vamp2 | -0.020 |
| fut9d | 0.046 | dpysl2b | -0.020 |
| lye | 0.046 | mbd3b | -0.020 |
| zgc:163030 | 0.046 | zgc:153409 | -0.020 |
| elovl7b | 0.045 | atp6v1e1b | -0.019 |




