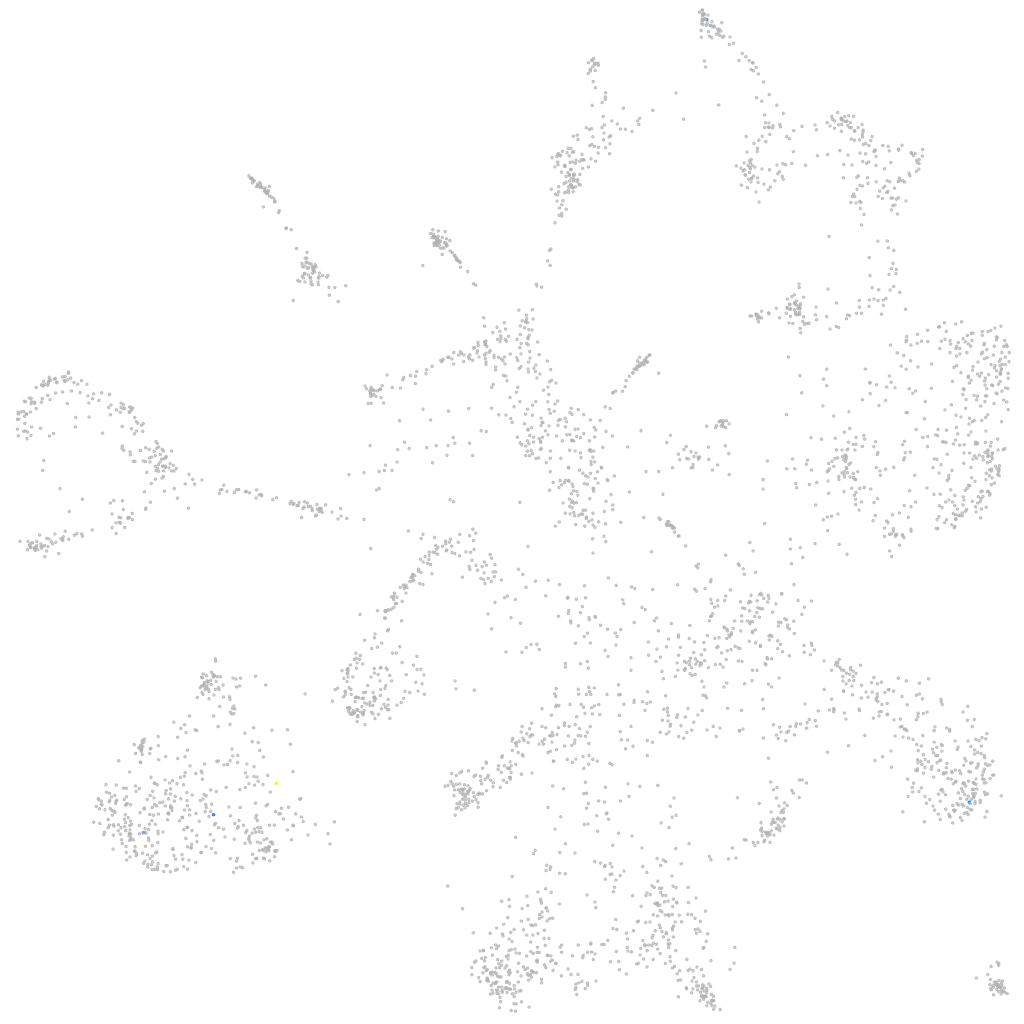
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| anpepa | 0.774 | rplp2 | -0.054 |
| CDCP1 | 0.531 | tmsb4x | -0.033 |
| si:dkey-45h7.1 | 0.449 | sparc | -0.033 |
| prdm1b | 0.326 | sept10 | -0.031 |
| XLOC-028703 | 0.291 | zgc:153867 | -0.029 |
| lingo4b | 0.290 | jun | -0.028 |
| CU076099.1 | 0.288 | fthl27 | -0.027 |
| sept4a | 0.286 | junba | -0.026 |
| c1galt1a | 0.265 | zgc:162730 | -0.026 |
| CR936482.1 | 0.249 | arpc3 | -0.025 |
| igsf9b | 0.239 | BX088707.3 | -0.025 |
| XLOC-032722 | 0.230 | GCA | -0.025 |
| otx2b | 0.230 | sec61g | -0.025 |
| CT990618.1 | 0.230 | capns1b | -0.024 |
| BX530077.1 | 0.228 | mfap2 | -0.024 |
| neurog1 | 0.226 | jpt1b | -0.024 |
| rad52 | 0.211 | fstl1b | -0.024 |
| nectin1a | 0.199 | ier2b | -0.024 |
| cep68 | 0.187 | btg2 | -0.024 |
| gng8 | 0.186 | si:dkey-7j14.6 | -0.023 |
| xrcc3 | 0.185 | oaz1b | -0.023 |
| fut10 | 0.183 | ctsla | -0.023 |
| CABZ01071972.1 | 0.181 | tegt | -0.022 |
| cd36 | 0.180 | rpl22l1 | -0.022 |
| tnfsf13b | 0.180 | ier2a | -0.022 |
| crx | 0.179 | socs3a | -0.022 |
| si:ch73-144d13.7 | 0.174 | id3 | -0.022 |
| asb6 | 0.172 | sdc4 | -0.022 |
| fbxo7 | 0.169 | col1a2 | -0.022 |
| si:dkey-238o13.4 | 0.161 | eif2s1b | -0.022 |
| kif20bb | 0.161 | eif3f | -0.021 |
| CABZ01085139.1 | 0.159 | eef1da | -0.021 |
| hexa | 0.159 | COX3 | -0.021 |
| idi1 | 0.157 | si:ch1073-291c23.2 | -0.021 |
| gadd45gb.1 | 0.155 | foxf1 | -0.021 |