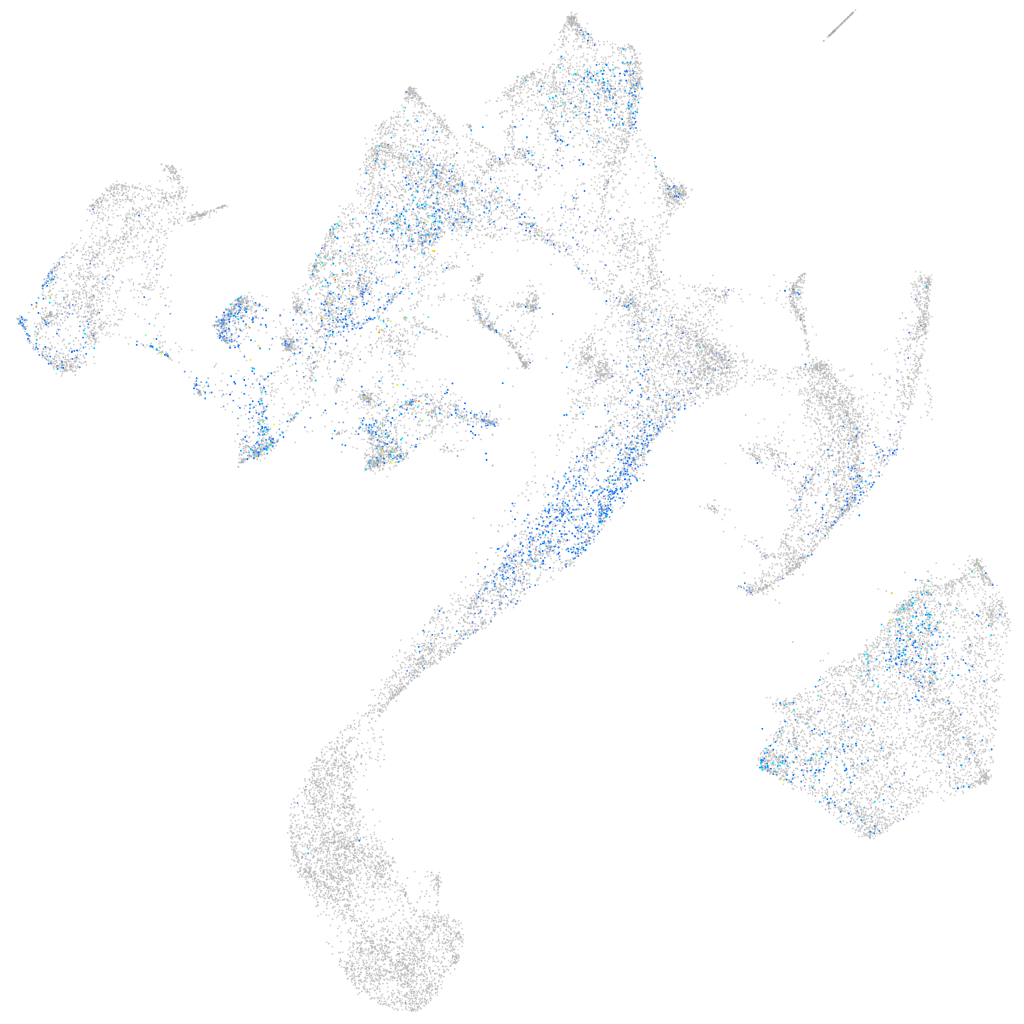
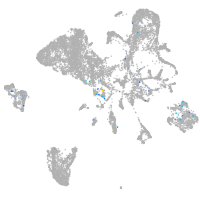
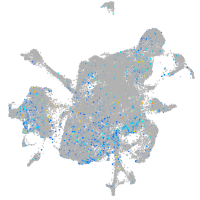
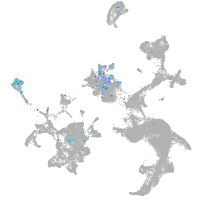
activator of transcription and developmental regulator AUTS2 b
ZFIN 
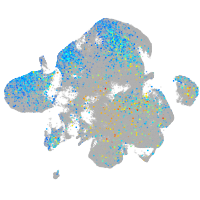
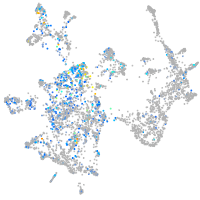
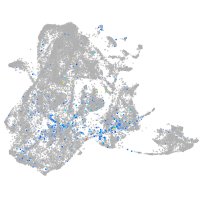
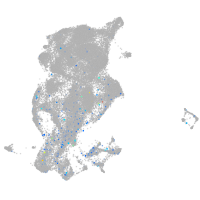
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| myog | 0.175 | tnni2a.4 | -0.099 |
| jam2a | 0.150 | myoz1b | -0.098 |
| efhd1 | 0.148 | mylpfb | -0.097 |
| mafbb | 0.130 | ryr3 | -0.090 |
| thrap3a | 0.128 | pvalb2 | -0.089 |
| si:ch1073-268j14.1 | 0.120 | pvalb1 | -0.088 |
| fam107b | 0.119 | actn3a | -0.087 |
| mafaa | 0.116 | myhz1.3 | -0.085 |
| megf10 | 0.115 | smyd1a | -0.084 |
| sh3d19 | 0.114 | slc25a4 | -0.083 |
| mymk | 0.114 | tpma | -0.083 |
| tmsb | 0.112 | casq1a | -0.081 |
| rbm24a | 0.108 | igfn1.1 | -0.080 |
| phlda2 | 0.105 | CABZ01061524.1 | -0.079 |
| myhb | 0.105 | myhz1.1 | -0.078 |
| mef2d | 0.104 | myoz2a | -0.078 |
| cxxc5a | 0.099 | tmod4 | -0.077 |
| aktip | 0.099 | acta1b | -0.076 |
| fbxl22 | 0.098 | myom2a | -0.076 |
| tuba8l3 | 0.097 | XLOC-005350 | -0.074 |
| mir133c | 0.095 | si:ch73-366l1.5 | -0.073 |
| mapre1a | 0.095 | tgm2a | -0.073 |
| tbx1 | 0.094 | mylz3 | -0.072 |
| traf4a | 0.093 | XLOC-006515 | -0.072 |
| lmnb1 | 0.092 | si:ch73-367p23.2 | -0.072 |
| fhl2a | 0.090 | si:ch211-255p10.3 | -0.072 |
| ppp1r14bb | 0.090 | ldha | -0.071 |
| plekho1b | 0.089 | myhz1.2 | -0.070 |
| pkig | 0.089 | pgam2 | -0.069 |
| CT573344.1 | 0.089 | myom1a | -0.069 |
| tcf12 | 0.089 | egfl6 | -0.068 |
| myca | 0.089 | XLOC-001975 | -0.068 |
| s1pr5a | 0.089 | hhatla | -0.068 |
| tnni2a.1 | 0.088 | hsc70 | -0.068 |
| cdkn1bb | 0.088 | igfn1.3 | -0.068 |