"ATRX chromatin remodeler, like"
ZFIN 
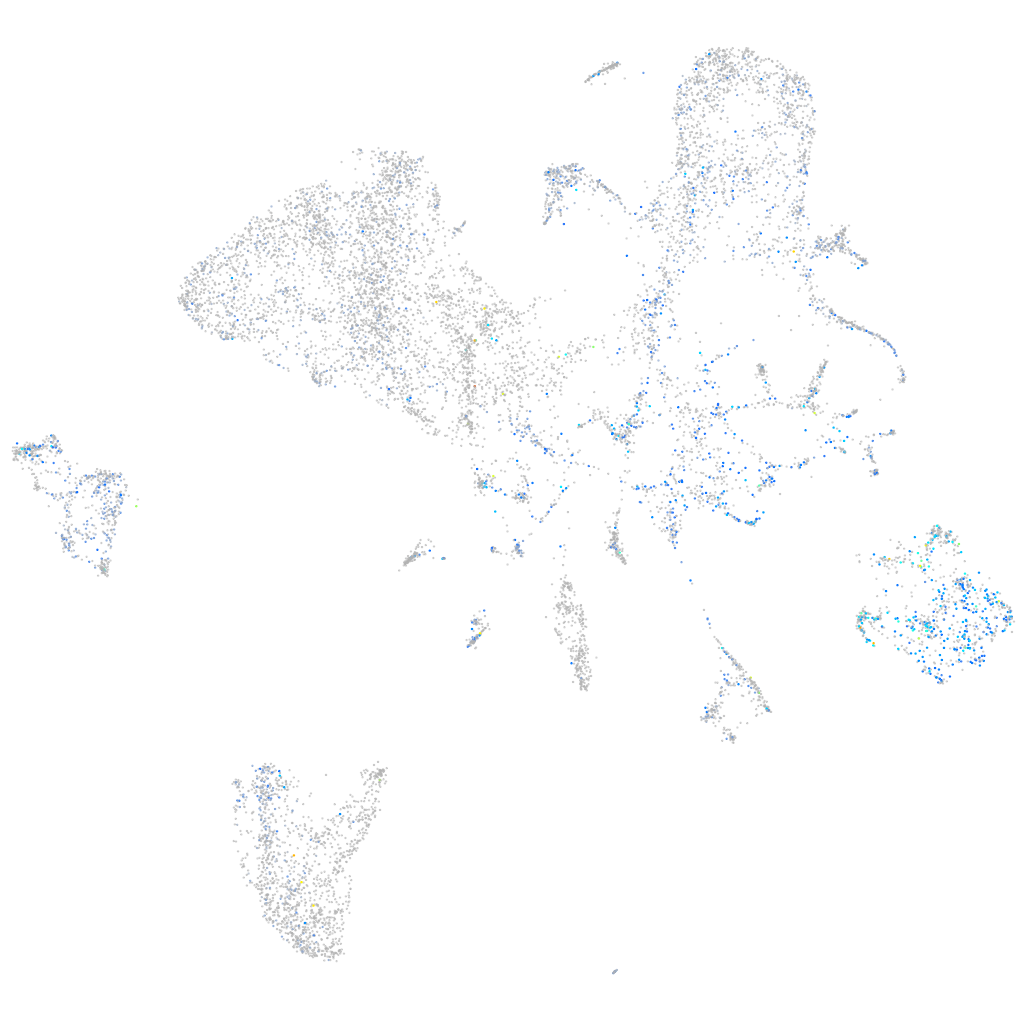
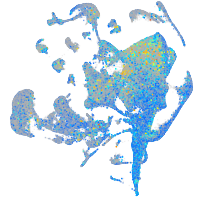
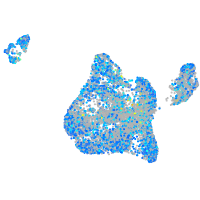
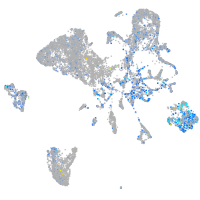
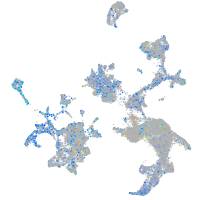
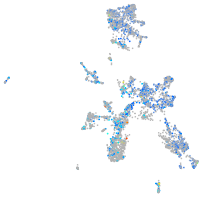
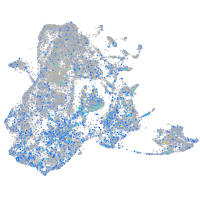
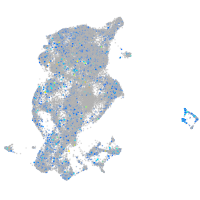
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hnrnpub | 0.289 | gamt | -0.218 |
| hmga1a | 0.284 | gapdh | -0.207 |
| hnrnpa1a | 0.284 | rpl37 | -0.203 |
| marcksb | 0.284 | ahcy | -0.198 |
| nucks1a | 0.282 | gatm | -0.197 |
| marcksl1b | 0.280 | rps10 | -0.192 |
| hnrnpa1b | 0.278 | bhmt | -0.189 |
| acin1a | 0.278 | apoa1b | -0.181 |
| si:ch211-152c2.3 | 0.277 | gpx4a | -0.180 |
| ilf3b | 0.275 | zgc:114188 | -0.176 |
| smarca4a | 0.274 | zgc:92744 | -0.169 |
| smc1al | 0.273 | mat1a | -0.169 |
| cx43.4 | 0.273 | apoa4b.1 | -0.168 |
| hspb1 | 0.273 | suclg1 | -0.168 |
| seta | 0.272 | fbp1b | -0.166 |
| syncrip | 0.271 | apoa2 | -0.165 |
| snrnp70 | 0.271 | afp4 | -0.163 |
| akap12b | 0.270 | apoc2 | -0.163 |
| khdrbs1a | 0.270 | nupr1b | -0.162 |
| cbx1a | 0.268 | agxtb | -0.160 |
| hnrnpabb | 0.268 | eno3 | -0.160 |
| hnrnpm | 0.266 | rps17 | -0.159 |
| chaf1a | 0.265 | pnp4b | -0.157 |
| hdac1 | 0.265 | pklr | -0.157 |
| top1l | 0.265 | glud1b | -0.156 |
| tardbp | 0.265 | scp2a | -0.156 |
| cirbpa | 0.264 | rbp4 | -0.154 |
| hnrnpaba | 0.264 | grhprb | -0.153 |
| hmgb2a | 0.263 | aldob | -0.151 |
| ppig | 0.262 | zgc:123103 | -0.151 |
| brd3a | 0.260 | sod1 | -0.151 |
| anp32a | 0.259 | abat | -0.150 |
| si:ch73-1a9.3 | 0.258 | apobb.1 | -0.149 |
| hnrnpa0a | 0.258 | apom | -0.148 |
| hnrnpa0b | 0.257 | fetub | -0.147 |