ATPase H+ transporting V1 subunit G1
ZFIN 
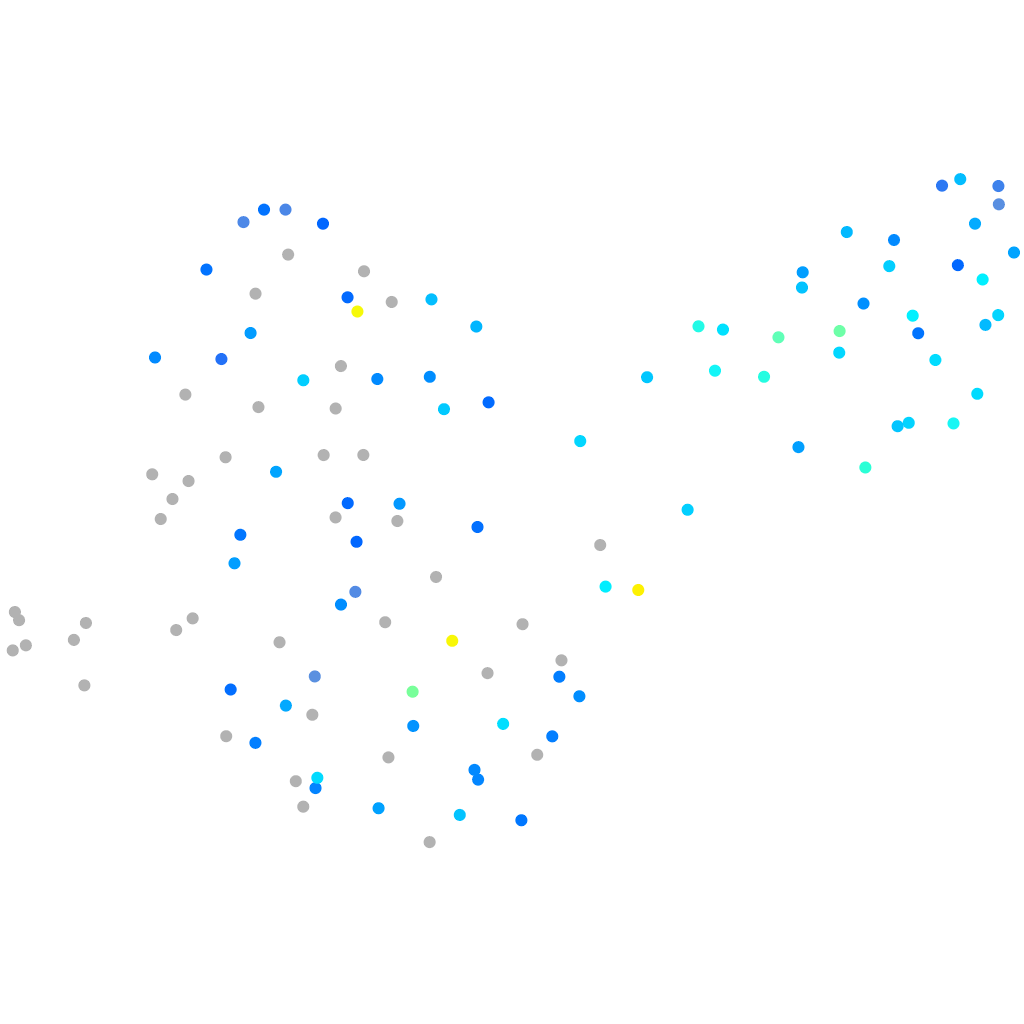
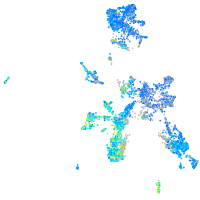
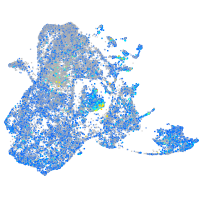
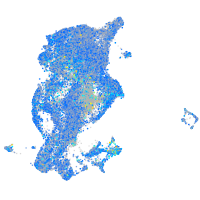
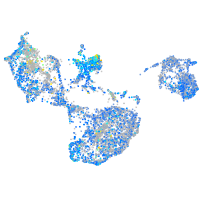
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cox6a1 | 0.682 | nop58 | -0.609 |
| rps17 | 0.648 | h1m | -0.574 |
| rnasekb | 0.648 | marcksb | -0.565 |
| ndufa3 | 0.644 | ncl | -0.557 |
| si:ch1073-429i10.3.1 | 0.638 | npm1a | -0.549 |
| rpl28 | 0.628 | fbl | -0.541 |
| rpl37 | 0.627 | NC-002333.4 | -0.539 |
| rps10 | 0.622 | hnrnpa1b | -0.530 |
| cox8a | 0.620 | nucks1a | -0.514 |
| rps15a | 0.611 | ppig | -0.512 |
| ISCU (1 of many) | 0.611 | hdgfl2 | -0.504 |
| calm3a | 0.608 | cd2bp2 | -0.500 |
| ndufb8 | 0.605 | top1l | -0.496 |
| suclg1 | 0.602 | srrt | -0.492 |
| cct2 | 0.597 | hnrnpub | -0.483 |
| atp6ap2 | 0.589 | cbx1a | -0.483 |
| atp5l | 0.583 | acin1b | -0.480 |
| eef1db | 0.581 | cdk11b | -0.474 |
| eif1ad | 0.579 | snrpb2 | -0.473 |
| dap1b | 0.579 | nono | -0.472 |
| NC-002333.17 | 0.575 | sltm | -0.466 |
| rps14 | 0.574 | brd3a | -0.466 |
| lsm7 | 0.571 | si:ch211-161h7.4 | -0.459 |
| hmgn3 | 0.568 | nasp | -0.458 |
| si:dkey-28b4.7 | 0.565 | ccna2 | -0.453 |
| smdt1b | 0.563 | hnrnpa1a | -0.451 |
| fkbp1aa | 0.562 | tpx2 | -0.448 |
| rpl21 | 0.560 | sde2 | -0.447 |
| rps26l | 0.559 | AL935174.5 | -0.441 |
| pcbd1 | 0.559 | dlgap5 | -0.435 |
| golgb1 | 0.555 | si:ch73-281n10.2 | -0.426 |
| eif3k | 0.555 | dek | -0.426 |
| zgc:114188 | 0.554 | gdf3 | -0.423 |
| CABZ01072614.1 | 0.554 | akap12b | -0.422 |
| zgc:153409 | 0.553 | marcksl1b | -0.422 |