ATP synthase peripheral stalk subunit OSCP
ZFIN 
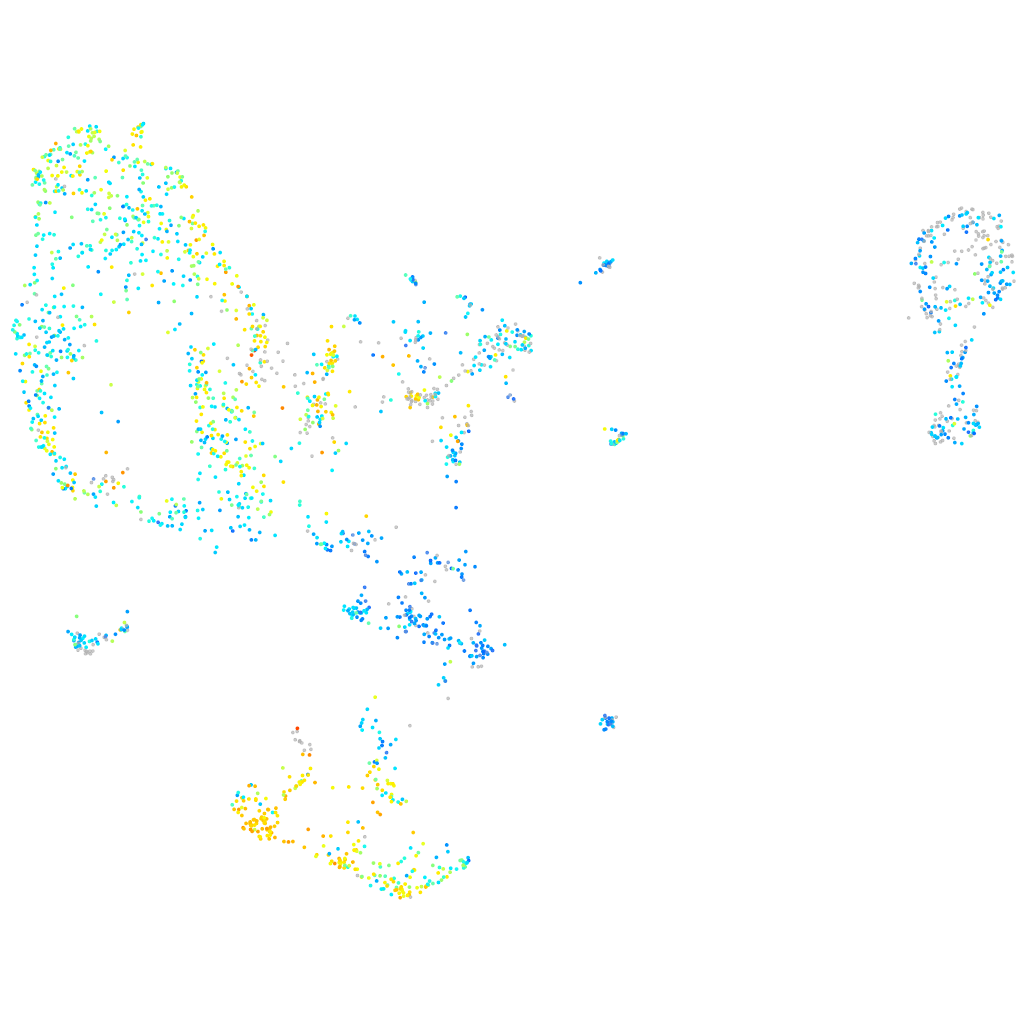
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| atp5f1b | 0.587 | hspb1 | -0.436 |
| atp5mc1 | 0.569 | marcksl1b | -0.423 |
| atp5l | 0.565 | wu:fb97g03 | -0.418 |
| ldhba | 0.557 | apoeb | -0.409 |
| atp5pd | 0.556 | marcksb | -0.408 |
| atp1b1a | 0.554 | hmgb2b | -0.399 |
| suclg1 | 0.549 | seta | -0.393 |
| cdh17 | 0.548 | si:ch73-1a9.3 | -0.393 |
| atp5fa1 | 0.546 | hnrnpabb | -0.389 |
| atp5mc3b | 0.544 | si:ch73-281n10.2 | -0.387 |
| atp5f1d | 0.535 | apoc1 | -0.387 |
| cox6a1 | 0.534 | nucks1a | -0.385 |
| si:ch211-139a5.9 | 0.531 | cx43.4 | -0.379 |
| cox5ab | 0.531 | setb | -0.373 |
| cox7a2a | 0.517 | anp32a | -0.369 |
| cox6b1 | 0.515 | hmga1a | -0.369 |
| eno3 | 0.514 | hnrnpaba | -0.363 |
| atp1a1a.4 | 0.512 | ilf3b | -0.361 |
| atp5pb | 0.508 | akap12b | -0.361 |
| mdh1aa | 0.506 | syncrip | -0.355 |
| COX5B | 0.505 | hmgb2a | -0.353 |
| atp5meb | 0.500 | hmgb1b | -0.350 |
| atp5pf | 0.499 | msna | -0.345 |
| atp5mf | 0.498 | zgc:110425 | -0.344 |
| atp5f1c | 0.497 | hmgn6 | -0.342 |
| ndufb8 | 0.494 | anp32e | -0.340 |
| si:ch1073-325m22.2 | 0.489 | cdx4 | -0.337 |
| cox8a | 0.489 | ptmab | -0.336 |
| mt-nd4 | 0.489 | cirbpa | -0.335 |
| uqcrq | 0.488 | h3f3d | -0.334 |
| cox6c | 0.488 | ncl | -0.332 |
| zgc:193541 | 0.487 | anp32b | -0.328 |
| si:dkey-16p21.8 | 0.484 | ewsr1a | -0.327 |
| ndufa4l | 0.482 | snrnp70 | -0.323 |
| atp5if1b | 0.481 | si:ch211-288g17.3 | -0.321 |