ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2a
ZFIN 
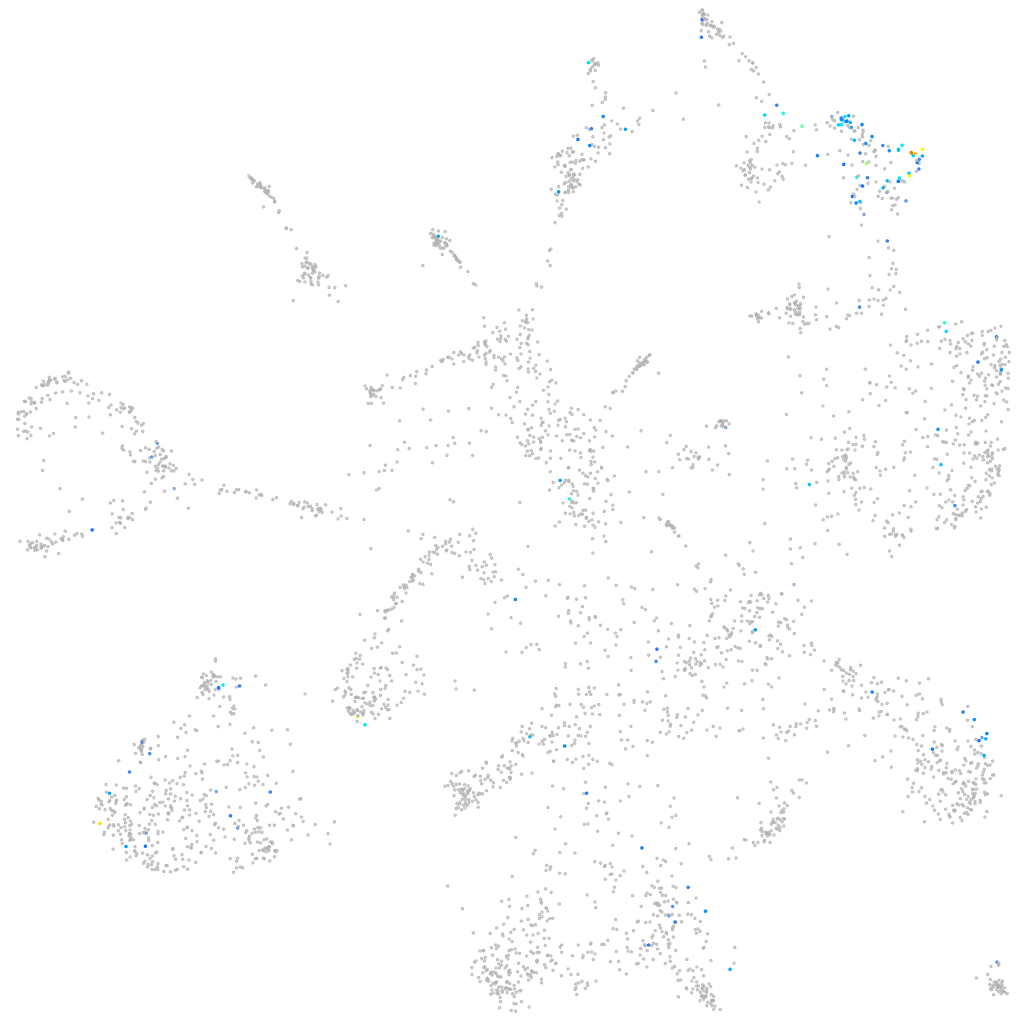
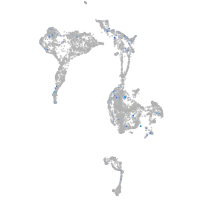
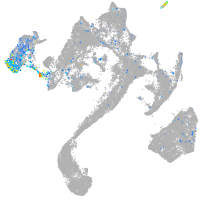
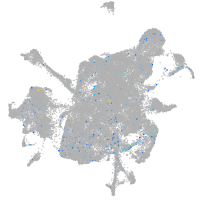
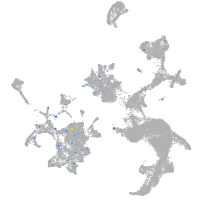
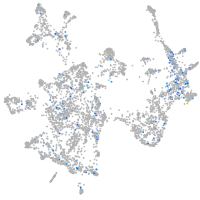
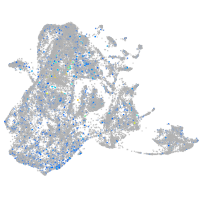
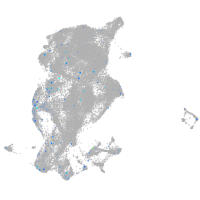
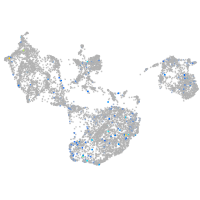
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tnni1b | 0.434 | XLOC-041870 | -0.105 |
| sfrp5 | 0.333 | tpm1 | -0.094 |
| cmlc1 | 0.331 | pmp22a | -0.089 |
| synpo2lb | 0.313 | BX088707.3 | -0.088 |
| mmel1 | 0.312 | XLOC-025423 | -0.086 |
| myh7bb | 0.297 | myl9a | -0.080 |
| alpi.1 | 0.293 | foxp4 | -0.074 |
| slc29a1b | 0.291 | cfd | -0.073 |
| lmod2a | 0.289 | gapdhs | -0.073 |
| tnnt2a | 0.288 | aoc2 | -0.068 |
| si:ch211-198m17.1 | 0.286 | rplp1 | -0.068 |
| bpifcl | 0.283 | acta2 | -0.066 |
| myh6 | 0.277 | XLOC-028370 | -0.066 |
| tbx5a | 0.275 | foxf1 | -0.065 |
| CABZ01075068.1 | 0.271 | aldh1a2 | -0.064 |
| XLOC-042222 | 0.268 | ifitm1 | -0.064 |
| actn2b | 0.256 | col1a2 | -0.062 |
| shisa2b | 0.242 | apoa2 | -0.061 |
| tnnc1a | 0.240 | vdac1 | -0.060 |
| mmp11a | 0.239 | crip1 | -0.060 |
| ryr2b | 0.239 | apoa1b | -0.060 |
| angpt1 | 0.233 | tagln | -0.058 |
| shisa3 | 0.232 | pbx3b | -0.058 |
| FITM1 (1 of many) | 0.227 | gpia | -0.057 |
| gata6 | 0.224 | lama4 | -0.057 |
| myl7 | 0.219 | pdgfrb | -0.057 |
| clec19a | 0.218 | myca | -0.057 |
| LOC103910903 | 0.213 | hlx1 | -0.057 |
| gata5 | 0.211 | BX322618.1 | -0.057 |
| ncs1a | 0.209 | nme2b.1 | -0.057 |
| bambib | 0.207 | prdx2 | -0.056 |
| nr0b2a | 0.207 | myl6 | -0.056 |
| smoc1 | 0.203 | nid2a | -0.056 |
| fabp11a | 0.203 | foxf2a | -0.056 |
| fbln2 | 0.202 | zgc:153704 | -0.056 |