ATPase phospholipid transporting 10A
ZFIN 
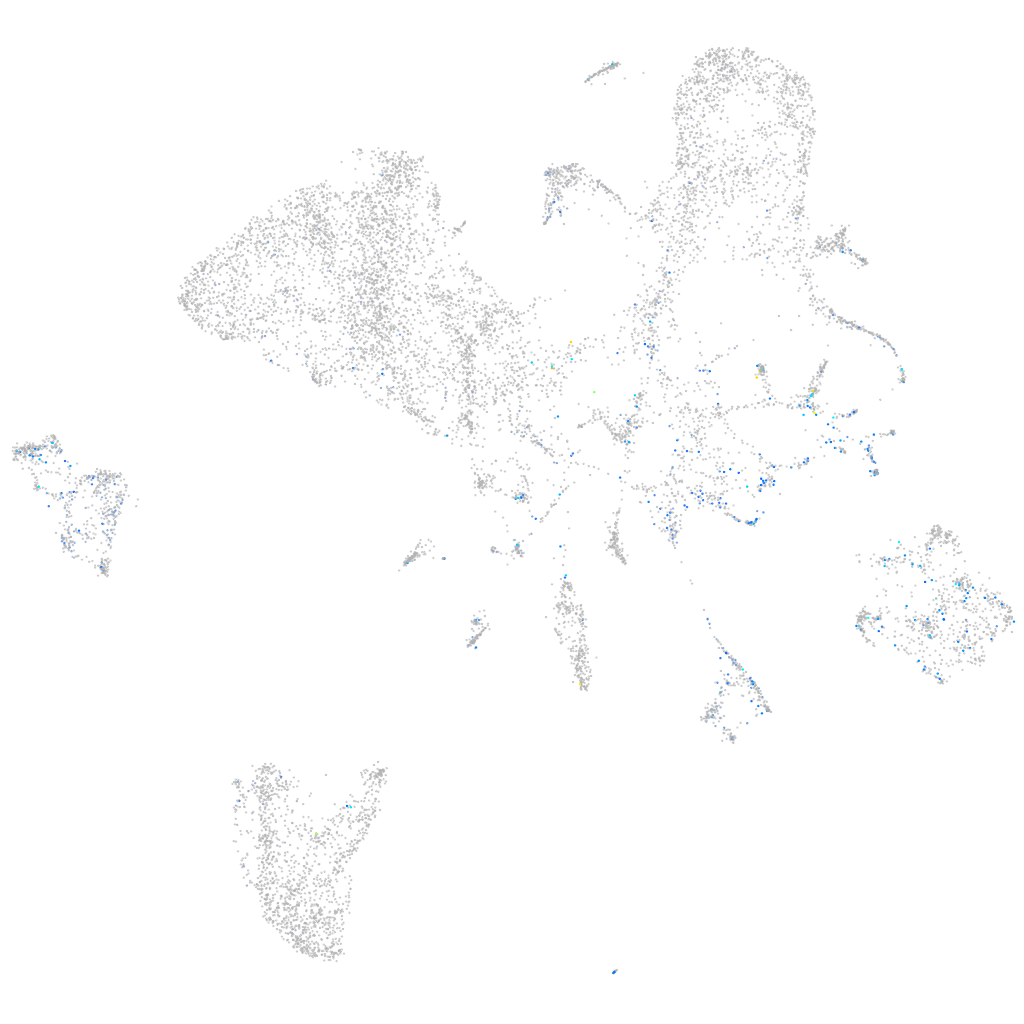
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:dkey-161j23.7 | 0.163 | gapdh | -0.177 |
| hmgn6 | 0.158 | gamt | -0.164 |
| si:ch211-222l21.1 | 0.150 | ahcy | -0.159 |
| si:ch73-1a9.3 | 0.145 | gatm | -0.145 |
| h3f3d | 0.145 | fbp1b | -0.144 |
| hmgb1a | 0.145 | apoa4b.1 | -0.142 |
| insm1a | 0.145 | gpx4a | -0.141 |
| h2afvb | 0.144 | mat1a | -0.135 |
| bcam | 0.141 | apoc2 | -0.133 |
| ubc | 0.141 | aldob | -0.133 |
| khdrbs1a | 0.138 | bhmt | -0.133 |
| zgc:92818 | 0.136 | apoa1b | -0.131 |
| hmgn2 | 0.136 | glud1b | -0.130 |
| lrrc75bb | 0.135 | eno3 | -0.130 |
| neurod1 | 0.135 | cx32.3 | -0.130 |
| epcam | 0.134 | scp2a | -0.127 |
| cirbpa | 0.133 | suclg1 | -0.126 |
| hmgb3a | 0.132 | dap | -0.126 |
| hmgb2a | 0.132 | afp4 | -0.125 |
| jpt1b | 0.132 | mdh1aa | -0.119 |
| ywhaz | 0.131 | gstt1a | -0.118 |
| ptmab | 0.131 | sult2st2 | -0.118 |
| hnrnpaba | 0.130 | abat | -0.118 |
| gnb1b | 0.130 | agxtb | -0.117 |
| si:dkey-42i9.4 | 0.130 | pnp4b | -0.117 |
| tspan7 | 0.130 | gcshb | -0.115 |
| hnrnpa0a | 0.130 | aldh6a1 | -0.115 |
| acin1a | 0.129 | nipsnap3a | -0.115 |
| syncrip | 0.129 | grhprb | -0.114 |
| pbx4 | 0.129 | rdh1 | -0.114 |
| anp32a | 0.129 | sod1 | -0.113 |
| nkx2.2a | 0.128 | tdo2a | -0.113 |
| csnk1a1 | 0.127 | apobb.1 | -0.113 |
| sumo3a | 0.127 | apoa2 | -0.112 |
| hmgn7 | 0.127 | aldh7a1 | -0.111 |