atonal bHLH transcription factor 7
ZFIN 
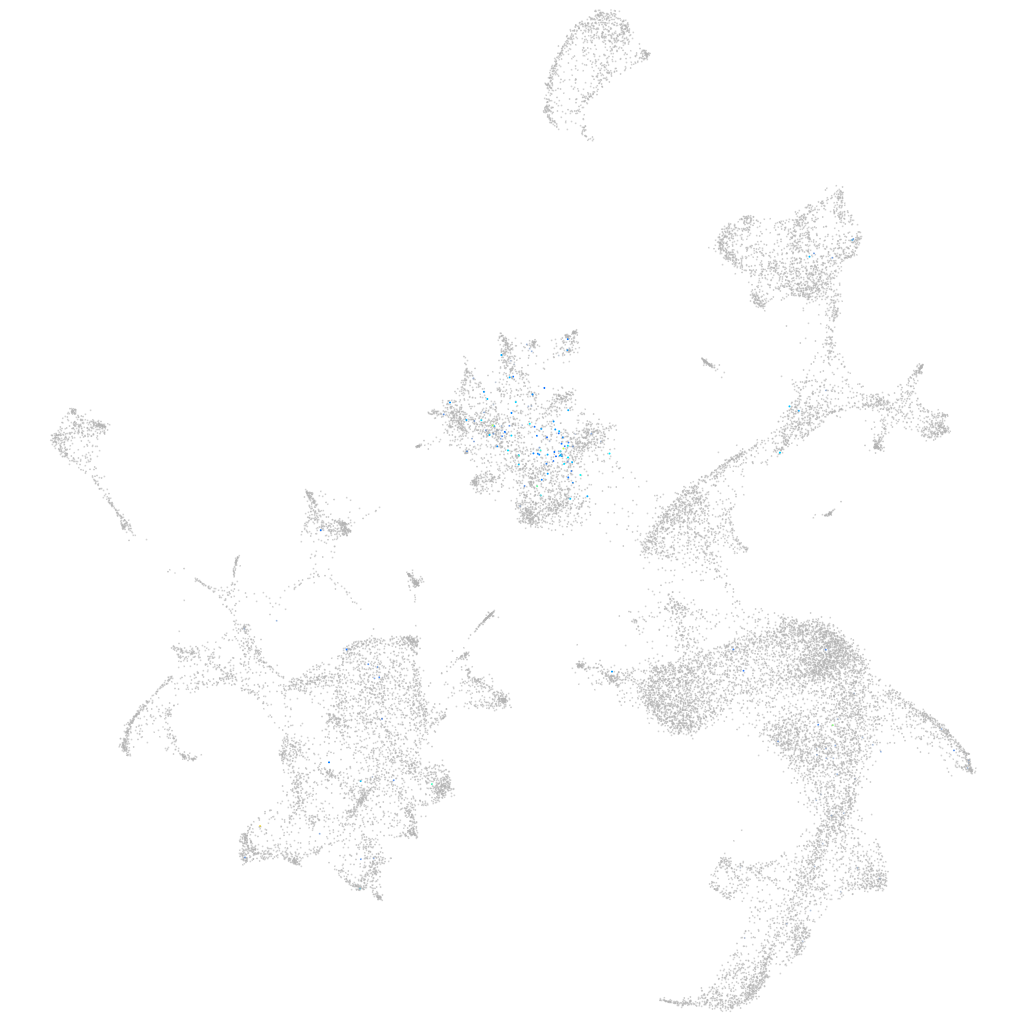
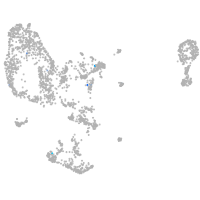
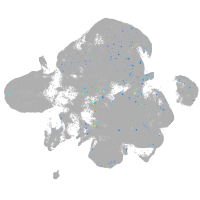
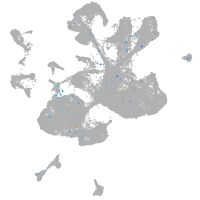
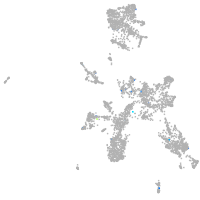
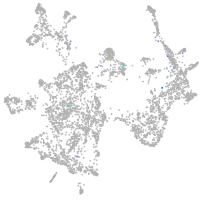
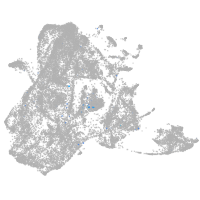
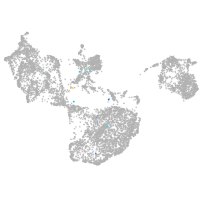
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| insm1a | 0.229 | mcl1a | -0.029 |
| hes2.1 | 0.205 | atp1b1a | -0.025 |
| hes2.2 | 0.182 | mob1a | -0.025 |
| foxn4 | 0.174 | lmo2 | -0.025 |
| im:7152348 | 0.170 | pfn1 | -0.024 |
| LOC103910639 | 0.148 | srgn | -0.022 |
| fabp7a | 0.147 | rac2 | -0.021 |
| pimr12 | 0.137 | znfl2a | -0.021 |
| gadd45gb.1 | 0.137 | dnase1l4.2 | -0.021 |
| hes6 | 0.136 | dusp5 | -0.020 |
| neurod4 | 0.135 | itm2bb | -0.020 |
| foxg1b | 0.131 | mbnl1 | -0.020 |
| cadm3 | 0.123 | hhex | -0.020 |
| LOC100537384 | 0.123 | si:ch73-248e21.7 | -0.019 |
| CU634008.1 | 0.122 | wasa | -0.019 |
| CABZ01113369.1 | 0.120 | coro1a | -0.019 |
| pax6a | 0.119 | ccr9a | -0.019 |
| BX294661.1 | 0.115 | rgs13 | -0.019 |
| kcnk3a | 0.115 | cebpb | -0.018 |
| si:dkeyp-34c12.1 | 0.115 | laptm5 | -0.018 |
| tuba1a | 0.113 | nrros | -0.018 |
| BX470218.1 | 0.112 | hcls1 | -0.018 |
| LOC108182767 | 0.110 | parvg | -0.018 |
| BX323543.4 | 0.106 | nr4a1 | -0.018 |
| LOC793648 | 0.105 | hmbsa | -0.018 |
| AL954695.3 | 0.105 | mafbb | -0.018 |
| CU459147.1 | 0.103 | arhgdig | -0.018 |
| hmgb1b | 0.102 | snx9b | -0.017 |
| XLOC-018654 | 0.101 | DHDH | -0.017 |
| elavl3 | 0.100 | si:dkey-262k9.4 | -0.017 |
| tfap2d | 0.100 | mhc1zba | -0.017 |
| epb41a | 0.098 | adam10a | -0.017 |
| mdkb | 0.097 | dub | -0.017 |
| ncam1a | 0.096 | wu:fb97g03 | -0.017 |
| six6b | 0.095 | spi1b | -0.017 |