autophagy related 3
ZFIN 
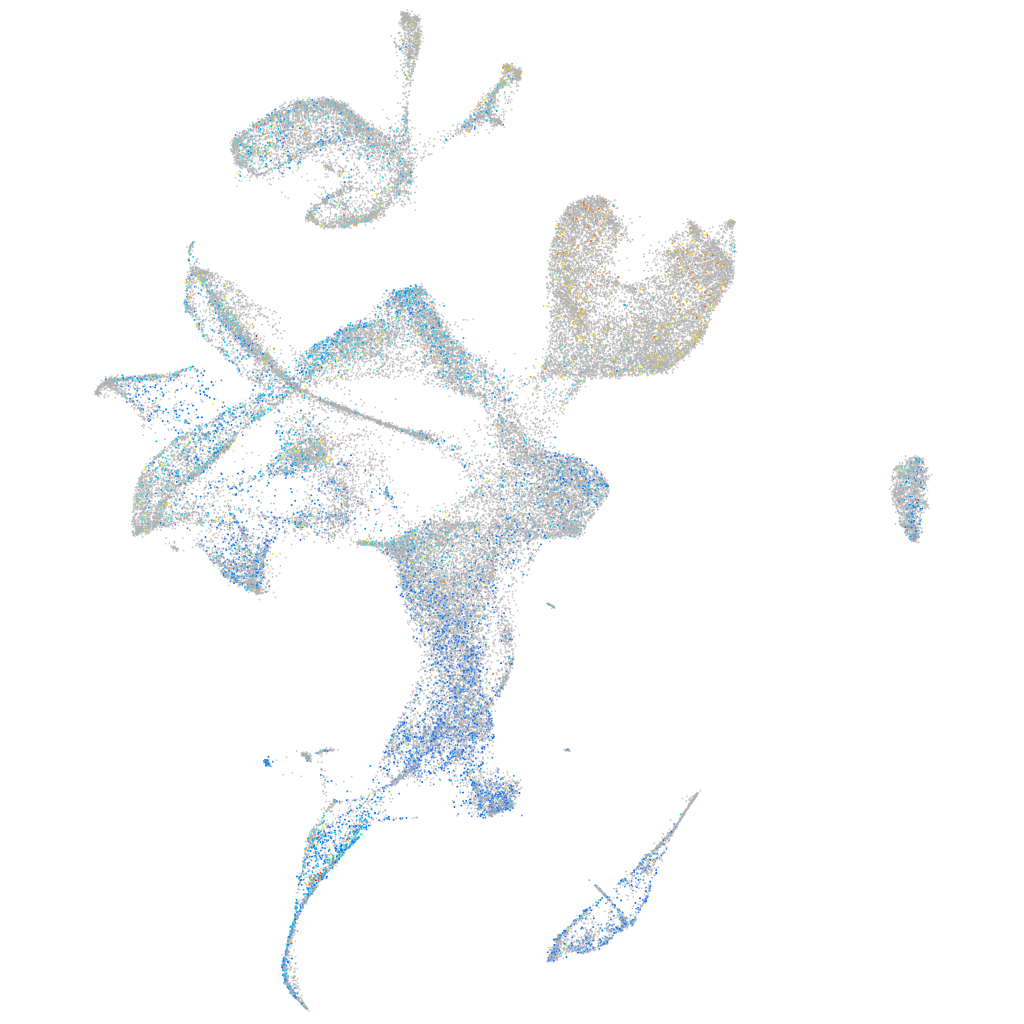
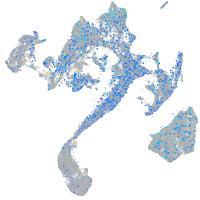
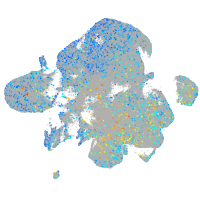
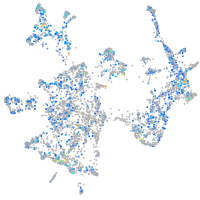
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tspan36 | 0.122 | ptmab | -0.075 |
| gpr143 | 0.120 | gpm6aa | -0.065 |
| qdpra | 0.117 | nova2 | -0.063 |
| slc45a2 | 0.117 | ptmaa | -0.057 |
| rab38 | 0.113 | mdkb | -0.057 |
| dct | 0.113 | si:ch73-1a9.3 | -0.056 |
| pcbd1 | 0.110 | scrt2 | -0.055 |
| pmela | 0.110 | hmgb1a | -0.051 |
| mitfa | 0.109 | si:ch211-222l21.1 | -0.047 |
| gstp1 | 0.108 | neurod4 | -0.047 |
| zgc:110591 | 0.107 | tp53inp2 | -0.045 |
| tyrp1b | 0.106 | tuba1a | -0.044 |
| agtrap | 0.103 | hes2.2 | -0.044 |
| pmelb | 0.101 | vsx1 | -0.043 |
| rab32a | 0.101 | bcl2l10 | -0.043 |
| pttg1ipb | 0.100 | im:7152348 | -0.042 |
| tyrp1a | 0.100 | myt1b | -0.042 |
| pnp4a | 0.100 | hes6 | -0.041 |
| FP085398.1 | 0.099 | rorb | -0.041 |
| rnaseka | 0.099 | LOC798783 | -0.039 |
| oca2 | 0.099 | foxn4 | -0.038 |
| syngr1a | 0.099 | hes2.1 | -0.038 |
| trpm1b | 0.098 | epb41a | -0.037 |
| slc24a5 | 0.098 | hmgn2 | -0.037 |
| fabp11b | 0.098 | btg1 | -0.037 |
| sparc | 0.097 | hunk | -0.037 |
| tspan10 | 0.097 | chd4a | -0.035 |
| tmem88b | 0.096 | dla | -0.035 |
| cx43 | 0.095 | pou3f1 | -0.035 |
| atox1 | 0.094 | insm1a | -0.035 |
| bace2 | 0.094 | myt1a | -0.035 |
| cd63 | 0.093 | CR383676.1 | -0.034 |
| zgc:110339 | 0.093 | tp53inp1 | -0.034 |
| tyr | 0.092 | dscaml1 | -0.034 |
| sdc4 | 0.092 | hmgb1b | -0.034 |