ATG12 autophagy related 12 homolog (S. cerevisiae)
ZFIN 
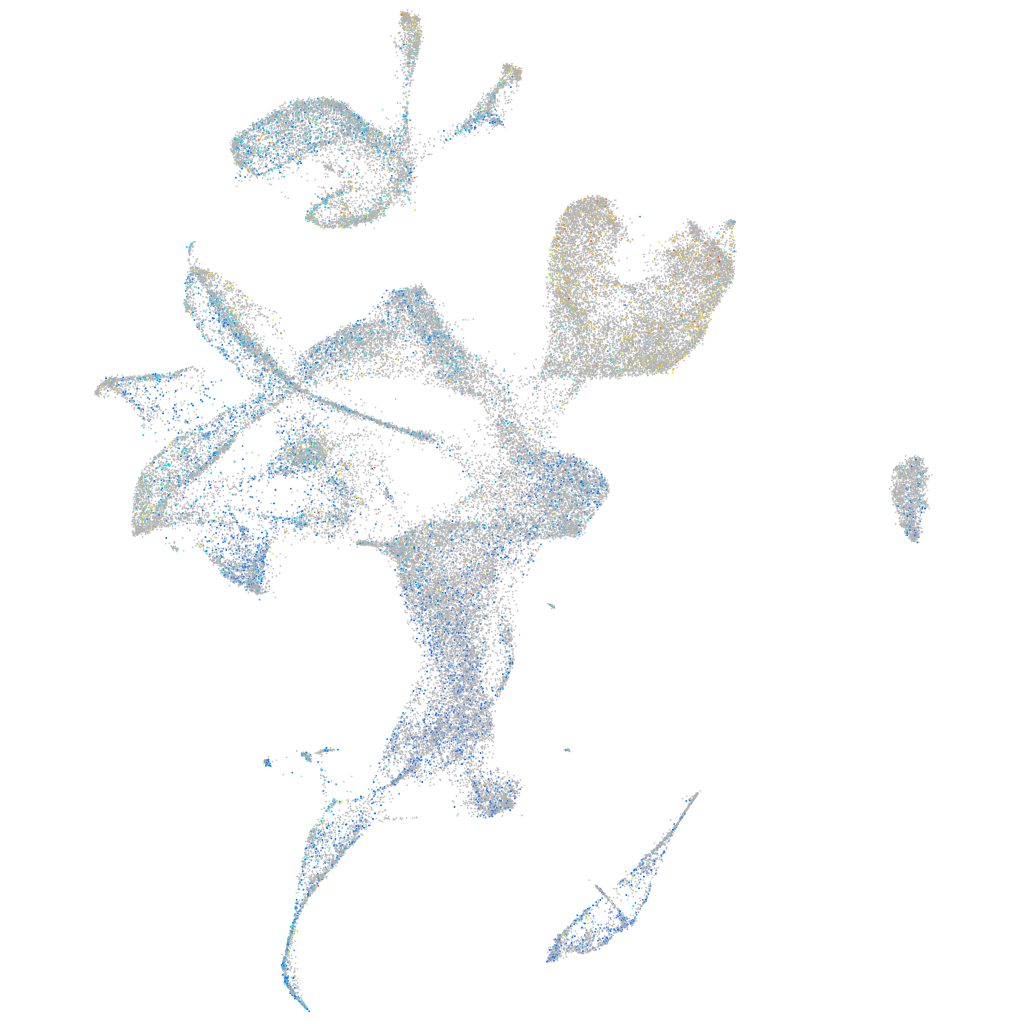
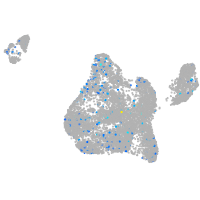
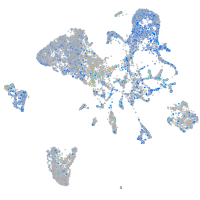
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| atp6v1e1b | 0.069 | si:ch211-222l21.1 | -0.051 |
| atp6v1g1 | 0.069 | hmga1a | -0.047 |
| ppdpfb | 0.062 | hmgb2a | -0.046 |
| calm2a | 0.061 | her15.1 | -0.042 |
| rnasekb | 0.058 | rrm1 | -0.041 |
| vat1 | 0.058 | pcna | -0.040 |
| igfbp7 | 0.058 | mcm7 | -0.039 |
| gng3 | 0.058 | nasp | -0.039 |
| brk1 | 0.058 | lig1 | -0.039 |
| gabarapl2 | 0.057 | mcm6 | -0.038 |
| atp5mc1 | 0.057 | mcm2 | -0.037 |
| calm1a | 0.056 | chaf1a | -0.037 |
| gpr143 | 0.056 | CABZ01005379.1 | -0.037 |
| sparc | 0.056 | ptmab | -0.036 |
| gapdhs | 0.056 | ccnd1 | -0.036 |
| atp6v0cb | 0.056 | hmgb2b | -0.036 |
| zgc:65894 | 0.055 | LOC798783 | -0.036 |
| atp6v1h | 0.055 | cdca7a | -0.036 |
| sncb | 0.055 | mcm5 | -0.035 |
| tyrp1b | 0.055 | mcm3 | -0.035 |
| id2a | 0.055 | hmgn2 | -0.034 |
| atp6ap2 | 0.054 | slbp | -0.034 |
| atp6v0b | 0.053 | XLOC-042899 | -0.034 |
| dct | 0.053 | mki67 | -0.034 |
| bace2 | 0.053 | dla | -0.033 |
| pttg1ipb | 0.053 | seta | -0.033 |
| mpc1 | 0.052 | mcm4 | -0.033 |
| ywhag2 | 0.052 | hes2.2 | -0.033 |
| cox7a2a | 0.052 | esco2 | -0.033 |
| syngr1a | 0.052 | cx43.4 | -0.032 |
| selenou1b | 0.052 | rpl15 | -0.032 |
| pmela | 0.052 | stmn1a | -0.032 |
| tspan36 | 0.052 | her4.2 | -0.031 |
| rlbp1b | 0.052 | rrm2 | -0.031 |
| h2afx1 | 0.051 | dek | -0.031 |