activating transcription factor 7 interacting protein
ZFIN 
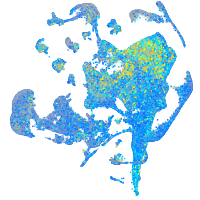
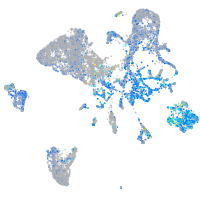
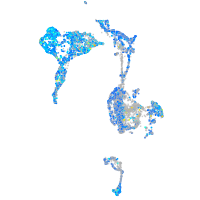
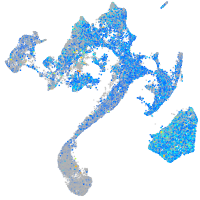
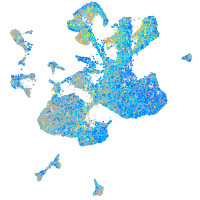
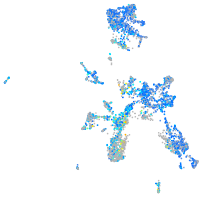
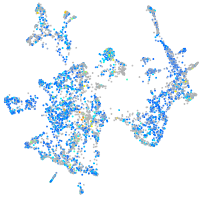
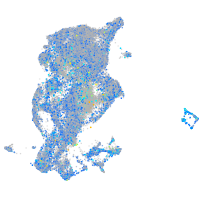
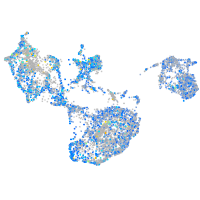
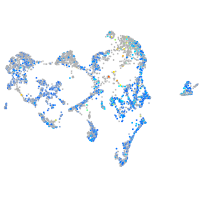
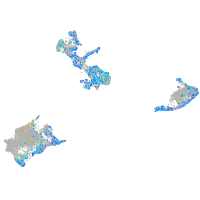
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hnrnpaba | 0.275 | eno3 | -0.233 |
| nucks1a | 0.258 | si:ch211-139a5.9 | -0.220 |
| hmgn6 | 0.253 | aldh6a1 | -0.217 |
| hnrnpa0b | 0.249 | grhprb | -0.210 |
| khdrbs1a | 0.246 | aldob | -0.210 |
| h3f3d | 0.245 | gapdh | -0.209 |
| hnrnpa0a | 0.245 | glud1b | -0.208 |
| h2afvb | 0.243 | fbp1b | -0.207 |
| ilf2 | 0.241 | sod1 | -0.205 |
| marcksb | 0.240 | dab2 | -0.204 |
| hnrnpabb | 0.240 | elovl1b | -0.202 |
| si:ch211-222l21.1 | 0.238 | pdzk1ip1 | -0.197 |
| cirbpb | 0.235 | aldh7a1 | -0.196 |
| cbx5 | 0.233 | cdaa | -0.196 |
| hmgb1b | 0.232 | gstp1 | -0.195 |
| setb | 0.227 | nit2 | -0.195 |
| h2afva | 0.226 | suclg1 | -0.194 |
| ptmab | 0.224 | dpydb | -0.193 |
| hmgb2b | 0.223 | lrp2a | -0.193 |
| sumo3a | 0.222 | cox6a2 | -0.193 |
| erh | 0.219 | upb1 | -0.193 |
| marcksl1b | 0.215 | gstr | -0.190 |
| hmgn2 | 0.213 | gcshb | -0.190 |
| tmeff1b | 0.213 | pklr | -0.189 |
| syncrip | 0.213 | si:dkey-33i11.9 | -0.189 |
| si:ch73-281n10.2 | 0.209 | cox6c | -0.188 |
| si:ch211-288g17.3 | 0.208 | si:dkey-16p21.8 | -0.187 |
| nono | 0.207 | dera | -0.187 |
| hmgb3a | 0.207 | hao1 | -0.187 |
| si:ch73-1a9.3 | 0.206 | ahcy | -0.186 |
| hmga1a | 0.205 | pdzk1 | -0.186 |
| ddx39ab | 0.205 | rgrb | -0.186 |
| ilf3b | 0.205 | pnp6 | -0.184 |
| hmgb2a | 0.204 | alpl | -0.184 |
| ctnnbip1 | 0.202 | gamt | -0.183 |