aspartate beta-hydroxylase domain containing 1
ZFIN 
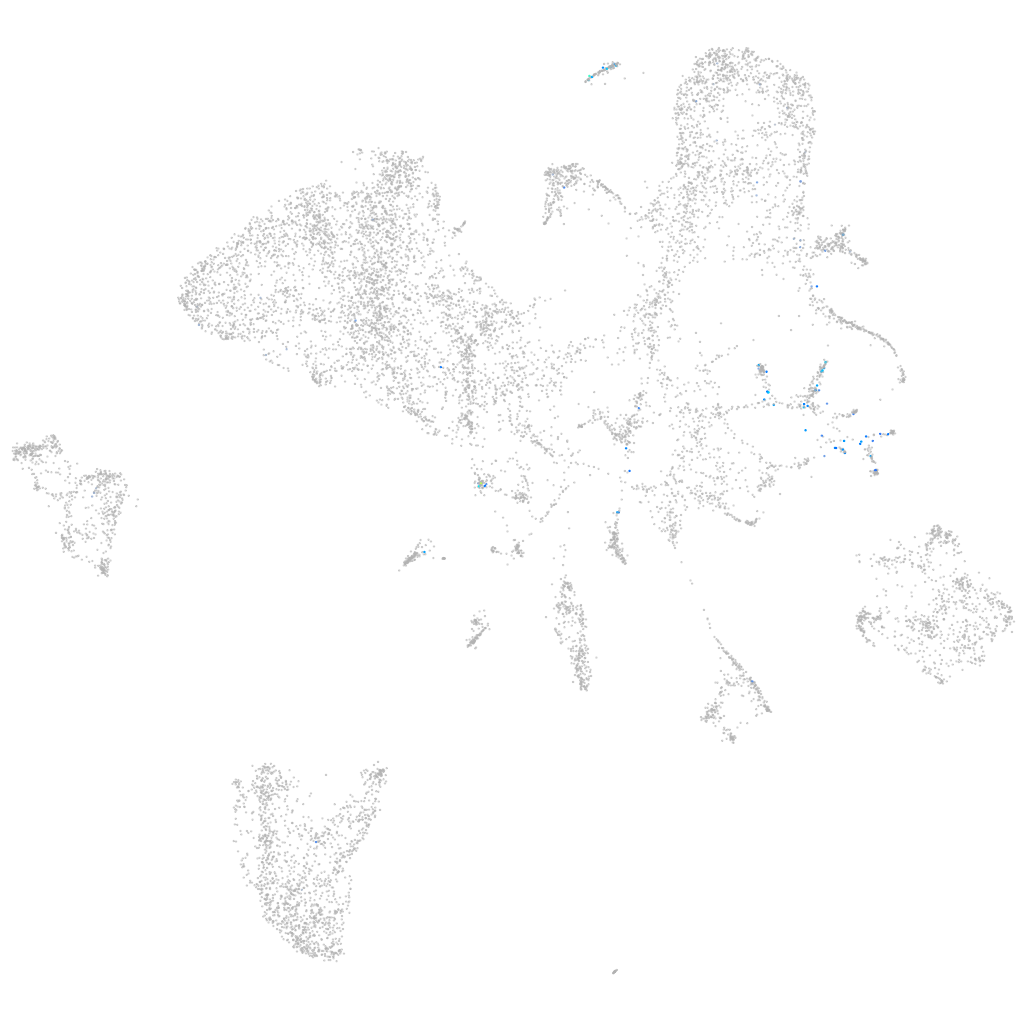
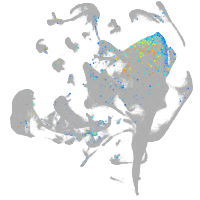
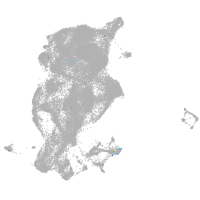
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.259 | gamt | -0.073 |
| or111-5 | 0.242 | gapdh | -0.072 |
| LOC108181470 | 0.228 | hdlbpa | -0.070 |
| ccdc177 | 0.226 | aldob | -0.068 |
| insm1b | 0.225 | ahcy | -0.067 |
| neurod1 | 0.223 | gatm | -0.065 |
| mir7a-1 | 0.219 | mat1a | -0.059 |
| tspan7b | 0.215 | cx32.3 | -0.058 |
| egr4 | 0.214 | fabp3 | -0.057 |
| pcsk1nl | 0.212 | rpl6 | -0.057 |
| zgc:101731 | 0.208 | bhmt | -0.057 |
| LOC103908906 | 0.204 | apoa1b | -0.054 |
| si:dkey-153k10.9 | 0.203 | fbp1b | -0.053 |
| insm1a | 0.203 | glud1b | -0.053 |
| hepacam2 | 0.202 | aqp12 | -0.053 |
| mir375-2 | 0.201 | si:dkey-16p21.8 | -0.053 |
| lysmd2 | 0.199 | aldh7a1 | -0.052 |
| phf1 | 0.199 | apoc2 | -0.052 |
| map1b | 0.198 | scp2a | -0.052 |
| BX890591.1 | 0.194 | agxtb | -0.052 |
| pax6b | 0.192 | aldh6a1 | -0.051 |
| c2cd4a | 0.190 | mgst1.2 | -0.051 |
| zgc:194578 | 0.189 | apoa4b.1 | -0.051 |
| vamp2 | 0.187 | hspd1 | -0.051 |
| id4 | 0.187 | gnmt | -0.051 |
| vwa5b2 | 0.186 | apoc1 | -0.051 |
| gdi1 | 0.185 | nupr1b | -0.051 |
| fev | 0.183 | slc16a1b | -0.050 |
| scgn | 0.183 | gstr | -0.050 |
| syt1a | 0.182 | agxta | -0.050 |
| glipr1b | 0.182 | afp4 | -0.050 |
| isl1 | 0.182 | suclg2 | -0.049 |
| scg2a | 0.182 | tfa | -0.048 |
| rnasekb | 0.181 | gstt1a | -0.048 |
| elovl1a | 0.180 | rgrb | -0.047 |