aristaless related homeobox b
ZFIN 
Other cell groups


all cells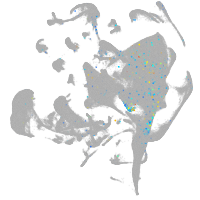 |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye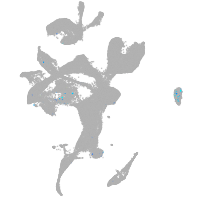 |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous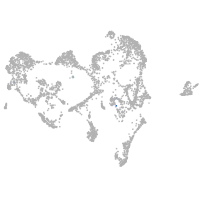 |
pigment cells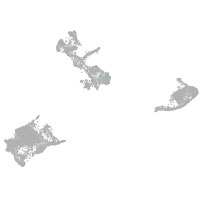 |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| arxa | 0.142 | rps10 | -0.046 |
| chrd | 0.137 | rpl37 | -0.045 |
| nitr11a | 0.118 | zgc:114188 | -0.043 |
| dlx2a | 0.114 | ppiab | -0.037 |
| sox19a | 0.112 | ptmaa | -0.034 |
| rnf135 | 0.106 | rps17 | -0.034 |
| dlx1a | 0.104 | h3f3a | -0.033 |
| hspb1 | 0.100 | si:ch1073-429i10.3.1 | -0.032 |
| dlx2b | 0.095 | COX3 | -0.031 |
| foxd5 | 0.093 | CR383676.1 | -0.029 |
| shisa2a | 0.093 | zgc:158463 | -0.028 |
| cyp26a1 | 0.089 | fabp7a | -0.028 |
| wnt7ba | 0.089 | cadm3 | -0.027 |
| LOC101885687 | 0.088 | hnrnpa0l | -0.027 |
| nkx2.9 | 0.085 | mt-nd1 | -0.026 |
| tdgf1 | 0.084 | mt-co2 | -0.025 |
| si:dkey-261m9.9.4 | 0.084 | pvalb1 | -0.025 |
| hesx1 | 0.084 | mt-atp6 | -0.025 |
| stm | 0.083 | gapdhs | -0.024 |
| si:ch211-152c2.3 | 0.083 | ckbb | -0.023 |
| efnb2b | 0.083 | cct2 | -0.023 |
| LOC101885266 | 0.079 | foxg1b | -0.023 |
| nr6a1a | 0.077 | actc1b | -0.023 |
| otx1 | 0.076 | rorb | -0.022 |
| BX004972.1 | 0.075 | neurod4 | -0.022 |
| LOC101885301 | 0.075 | gpm6aa | -0.022 |
| apip | 0.073 | hbae3 | -0.022 |
| olig3 | 0.073 | atp5mc1 | -0.021 |
| alcamb | 0.071 | gpm6ab | -0.021 |
| LOC103908936 | 0.071 | pvalb2 | -0.021 |
| foxb1b | 0.069 | NC-002333.17 | -0.021 |
| dlx5a | 0.069 | ahcy | -0.021 |
| XLOC-011211 | 0.069 | h3f3c | -0.020 |
| ISCU | 0.069 | atp5f1b | -0.020 |
| LOC110438343 | 0.068 | btg1 | -0.019 |




