Rho guanine nucleotide exchange factor (GEF) 7a
ZFIN 
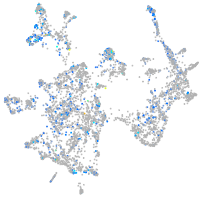
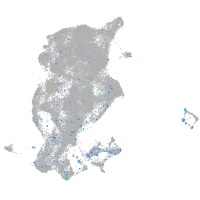
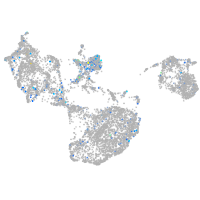
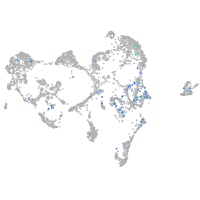
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| erc2.1 | 0.165 | gpr143 | -0.069 |
| nova2 | 0.161 | tspan36 | -0.064 |
| elavl3 | 0.142 | pcbd1 | -0.063 |
| CT990561.1 | 0.139 | smim29 | -0.062 |
| rnasekb | 0.137 | rnaseka | -0.059 |
| LOC110438343 | 0.136 | gstp1 | -0.059 |
| epb41a | 0.135 | qdpra | -0.059 |
| gpm6aa | 0.134 | tyr | -0.058 |
| celf3a | 0.133 | abracl | -0.056 |
| BX004770.1 | 0.132 | rps2 | -0.056 |
| hmgb3a | 0.131 | rab38 | -0.056 |
| nrxn1a | 0.131 | slc45a2 | -0.055 |
| stmn1b | 0.128 | cst14a.2 | -0.055 |
| hmgb1b | 0.127 | pts | -0.055 |
| rtn1a | 0.126 | zgc:165573 | -0.055 |
| UNC13A | 0.126 | ahnak | -0.054 |
| LOC110439819 | 0.125 | anxa2a | -0.054 |
| ifit10 | 0.125 | si:dkey-151g10.6 | -0.053 |
| caprin2 | 0.124 | si:dkey-204f11.64 | -0.053 |
| grxcr1a.1 | 0.124 | rpl36a | -0.052 |
| srrm4 | 0.124 | agtrap | -0.052 |
| cuedc1a | 0.123 | eef1a1l1 | -0.052 |
| gpr183a | 0.123 | impdh1b | -0.051 |
| slitrk1 | 0.119 | syngr1a | -0.051 |
| guca1e | 0.118 | rps24 | -0.051 |
| elavl4 | 0.117 | rpl12 | -0.051 |
| atp1a3a | 0.117 | zgc:110239 | -0.050 |
| tmem91 | 0.117 | slc37a2 | -0.049 |
| sox11b | 0.116 | pah | -0.049 |
| sypb | 0.116 | kita | -0.048 |
| si:ch211-195b15.8 | 0.115 | mtbl | -0.048 |
| lhfpl3 | 0.115 | bloc1s6 | -0.048 |
| si:ch211-222l21.1 | 0.115 | pnp4a | -0.048 |
| r3hdm1 | 0.115 | pttg1ipb | -0.047 |
| stmn4 | 0.115 | actb2 | -0.047 |