Rho guanine nucleotide exchange factor (GEF) 4
ZFIN 
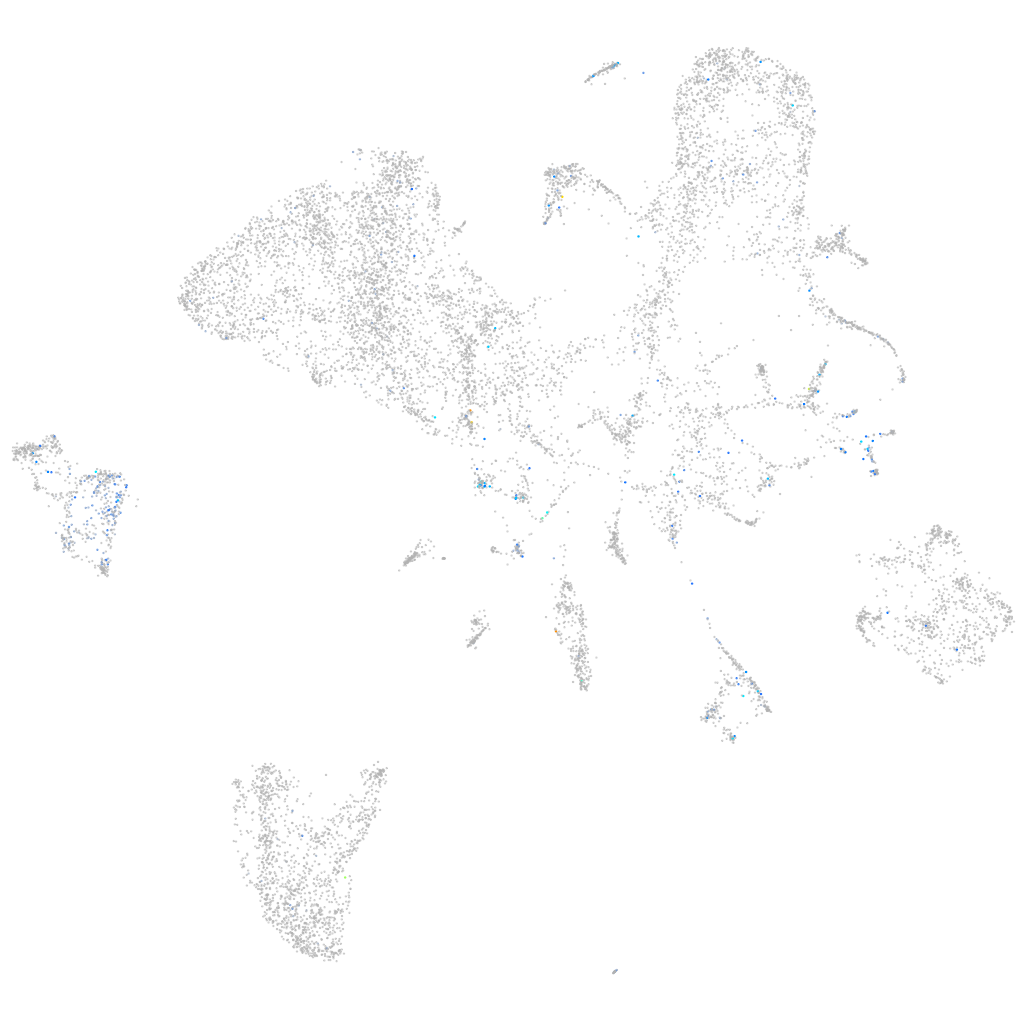
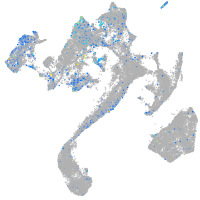
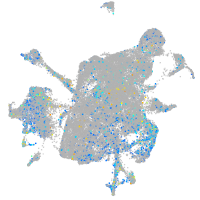
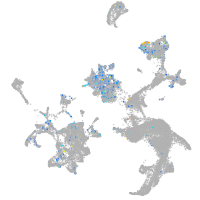
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| irf8 | 0.192 | gapdh | -0.107 |
| myo1f | 0.190 | gamt | -0.097 |
| LOC108180147 | 0.184 | ahcy | -0.087 |
| crygm2d20 | 0.172 | gatm | -0.085 |
| mettl11b | 0.166 | mat1a | -0.085 |
| XLOC-003396 | 0.164 | scp2a | -0.085 |
| tagapb | 0.162 | fbp1b | -0.081 |
| gpr31 | 0.161 | aqp12 | -0.080 |
| arhgef3l | 0.158 | dap | -0.079 |
| LOC108182795 | 0.157 | gpx4a | -0.077 |
| gapdhs | 0.155 | g6pca.2 | -0.077 |
| BX005419.1 | 0.151 | bhmt | -0.076 |
| ntng1a | 0.147 | gcshb | -0.076 |
| RF00560 | 0.146 | aldh7a1 | -0.075 |
| igfbp6b | 0.146 | aldob | -0.075 |
| flrt1b | 0.145 | apoc2 | -0.075 |
| CR450749.3 | 0.145 | nipsnap3a | -0.074 |
| si:ch211-281k19.2 | 0.145 | suclg1 | -0.074 |
| XLOC-012822 | 0.143 | gstt1a | -0.073 |
| rnasekb | 0.143 | apoa4b.1 | -0.073 |
| elavl4 | 0.142 | sult2st2 | -0.073 |
| pcdh2ab7 | 0.141 | cx32.3 | -0.073 |
| BX005448.1 | 0.140 | afp4 | -0.073 |
| XLOC-006525 | 0.140 | slc16a10 | -0.072 |
| sept5a | 0.139 | agxtb | -0.071 |
| XLOC-022188 | 0.138 | msrb2 | -0.071 |
| sost | 0.138 | rdh1 | -0.071 |
| CT955996.1 | 0.138 | tdo2a | -0.070 |
| ccl34b.1 | 0.135 | apoa1b | -0.070 |
| sostdc1a | 0.135 | hspe1 | -0.070 |
| enox1 | 0.134 | pnp4b | -0.070 |
| ttc12 | 0.133 | acmsd | -0.070 |
| ttc9b | 0.133 | cox8b | -0.070 |
| elavl3 | 0.133 | phyhd1 | -0.069 |
| spock1 | 0.131 | gstr | -0.069 |