Rho GDP dissociation inhibitor (GDI) alpha
ZFIN 
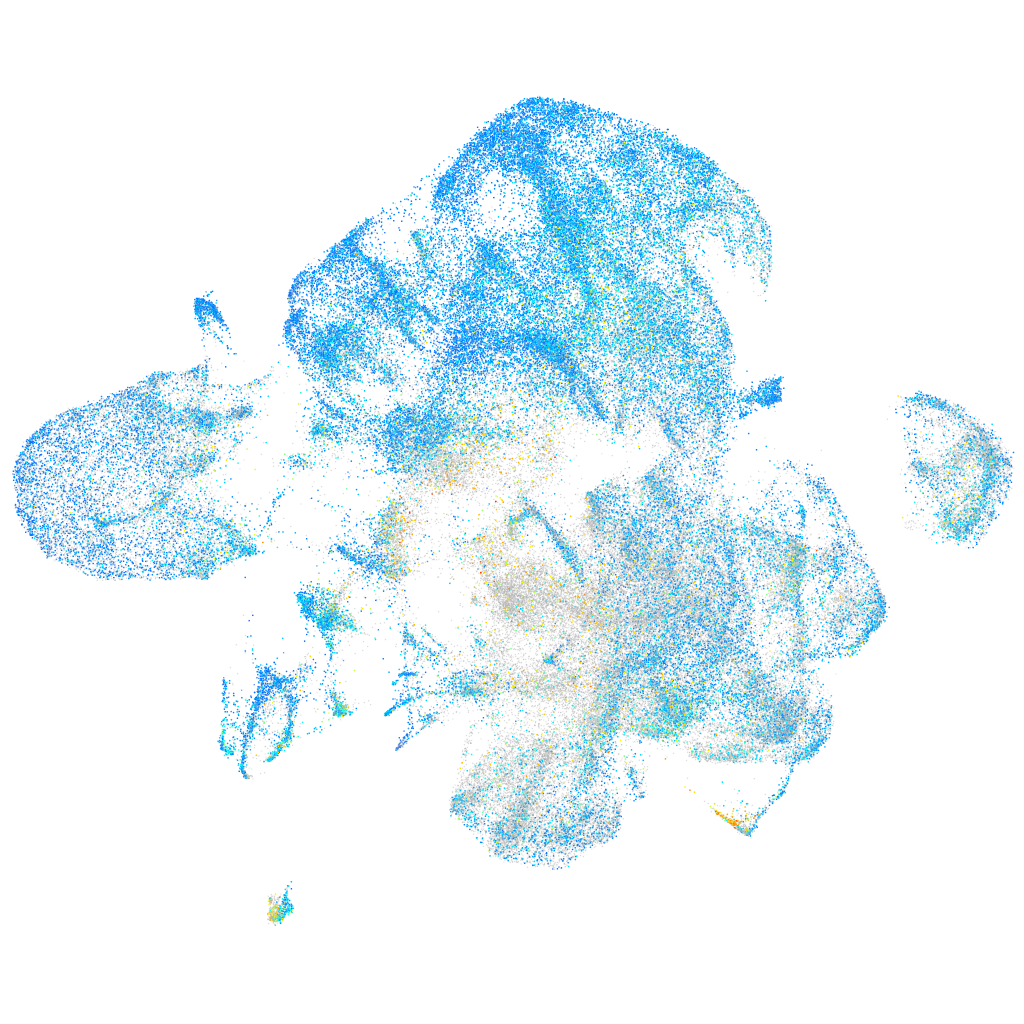
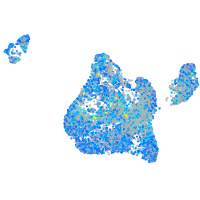
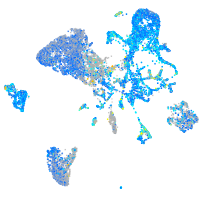
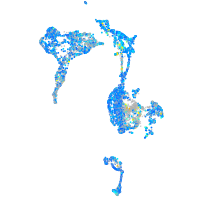
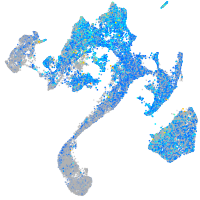
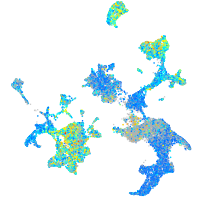
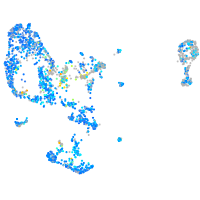
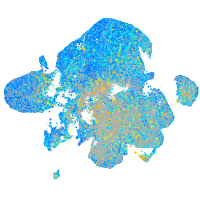
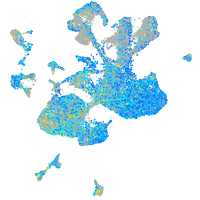
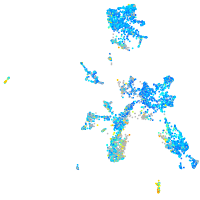
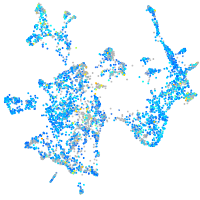
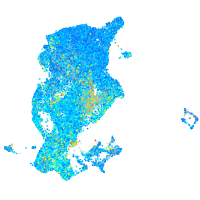
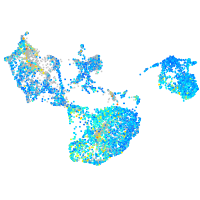
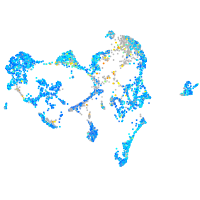
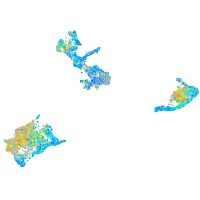
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| dut | 0.262 | stmn1b | -0.187 |
| tuba8l4 | 0.258 | gpm6ab | -0.183 |
| zgc:56493 | 0.256 | rtn1a | -0.181 |
| pcna | 0.251 | gpm6aa | -0.175 |
| hmgb2a | 0.245 | elavl3 | -0.172 |
| rrm1 | 0.240 | ckbb | -0.160 |
| ranbp1 | 0.232 | sncb | -0.154 |
| id1 | 0.231 | atp6v0cb | -0.151 |
| tuba8l | 0.230 | elavl4 | -0.151 |
| selenoh | 0.229 | gng3 | -0.150 |
| XLOC-003690 | 0.228 | rtn1b | -0.146 |
| ran | 0.226 | sox4a | -0.138 |
| stmn1a | 0.223 | zc4h2 | -0.135 |
| nutf2l | 0.219 | stx1b | -0.135 |
| mdka | 0.217 | fez1 | -0.133 |
| cldn5a | 0.217 | aplp1 | -0.132 |
| sox3 | 0.216 | nsg2 | -0.132 |
| mcm7 | 0.215 | rnasekb | -0.131 |
| anp32b | 0.215 | pbx3b | -0.131 |
| pa2g4b | 0.213 | ndrg4 | -0.128 |
| COX7A2 (1 of many) | 0.213 | tmem59l | -0.127 |
| her2 | 0.213 | myt1la | -0.126 |
| rrm2 | 0.212 | si:dkeyp-75h12.5 | -0.125 |
| XLOC-003689 | 0.212 | vamp2 | -0.125 |
| msna | 0.210 | rbfox1 | -0.124 |
| rpa2 | 0.209 | kdm6bb | -0.121 |
| cnbpa | 0.209 | tmsb | -0.121 |
| gfap | 0.209 | pvalb2 | -0.121 |
| rpa3 | 0.207 | adam22 | -0.121 |
| ptgr1 | 0.206 | pbx1a | -0.120 |
| ccna2 | 0.203 | zfhx3 | -0.120 |
| cks1b | 0.201 | pvalb1 | -0.120 |
| ccng1 | 0.200 | gabrb4 | -0.119 |
| vamp3 | 0.199 | cdk5r1b | -0.119 |
| fen1 | 0.198 | lhx1a | -0.118 |