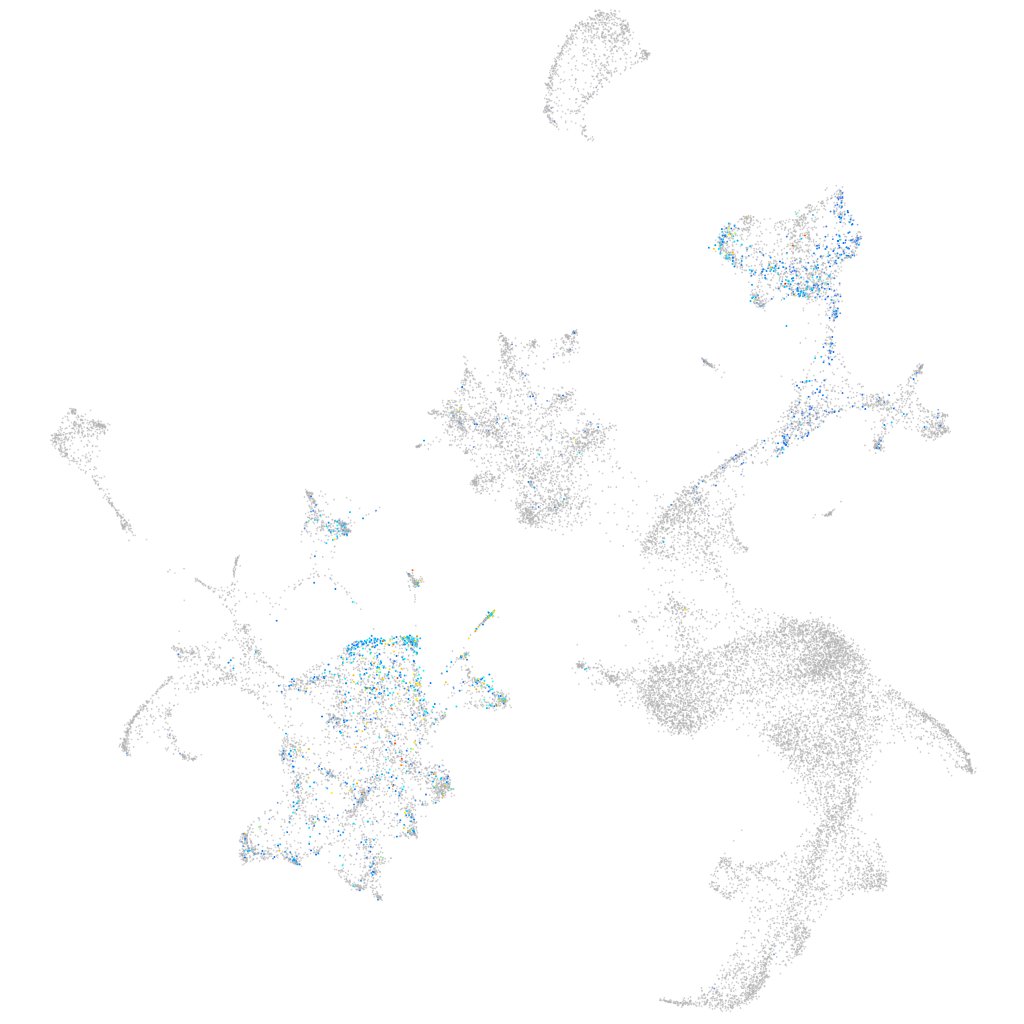
aquaporin 7
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm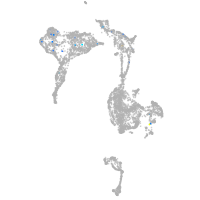 |
muscle 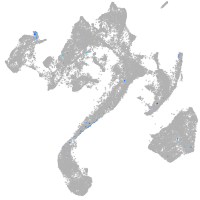 |
mural cells / non-skeletal muscle 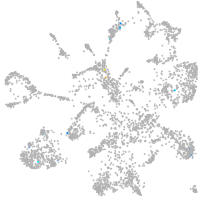 |
mesenchyme 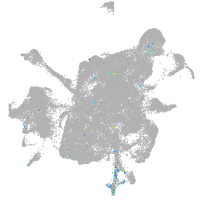 |
hematopoietic / vasculature 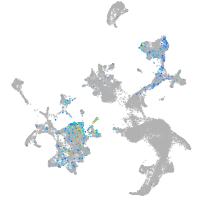 |
pronephros 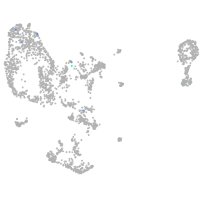 |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm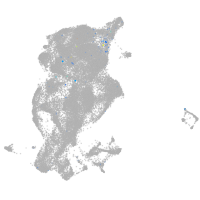 |
fin |
ionocytes / mucous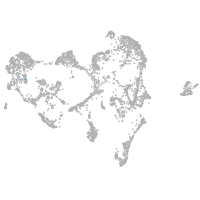 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mrc1a | 0.325 | hbae1.1 | -0.209 |
| lgmn | 0.308 | hbae1.3 | -0.202 |
| hexb | 0.306 | hbbe2 | -0.202 |
| lyve1b | 0.306 | hbae3 | -0.201 |
| zgc:110239 | 0.301 | hbbe1.3 | -0.197 |
| stab1 | 0.297 | hbbe1.1 | -0.194 |
| cpn1 | 0.295 | cahz | -0.189 |
| lamp2 | 0.285 | hemgn | -0.176 |
| ctsla | 0.285 | fth1a | -0.175 |
| si:dkey-28n18.9 | 0.284 | hbbe1.2 | -0.174 |
| stab2 | 0.283 | blvrb | -0.169 |
| dab2 | 0.283 | alas2 | -0.166 |
| cd63 | 0.275 | si:ch211-250g4.3 | -0.161 |
| npl | 0.273 | slc4a1a | -0.156 |
| gpr182 | 0.272 | nt5c2l1 | -0.154 |
| tpte | 0.268 | tspo | -0.154 |
| ctsh | 0.265 | zgc:163057 | -0.147 |
| gnpda1 | 0.262 | epb41b | -0.146 |
| cd81a | 0.262 | si:ch211-207c6.2 | -0.140 |
| fuca1.1 | 0.260 | creg1 | -0.140 |
| snx1a | 0.252 | nmt1b | -0.134 |
| npc2 | 0.252 | tmod4 | -0.130 |
| ctsz | 0.251 | prdx2 | -0.129 |
| fxyd6l | 0.249 | plac8l1 | -0.128 |
| mrc1b | 0.243 | zgc:56095 | -0.128 |
| ctsd | 0.242 | hbae5 | -0.117 |
| si:ch211-145b13.6 | 0.239 | sptb | -0.116 |
| dpp7 | 0.237 | tfr1a | -0.114 |
| zgc:110591 | 0.236 | rfesd | -0.111 |
| ifi30 | 0.232 | znfl2a | -0.110 |
| glula | 0.232 | uros | -0.109 |
| XLOC-016094 | 0.230 | add2 | -0.107 |
| junba | 0.230 | si:dkey-240n22.3 | -0.106 |
| snx2 | 0.229 | hbbe3 | -0.106 |
| ier2b | 0.229 | klf1 | -0.106 |