"amyloid beta (A4) precursor protein-binding, family A, member 1b"
ZFIN 
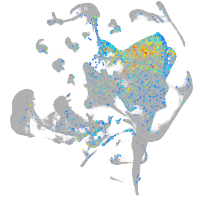
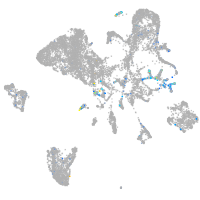
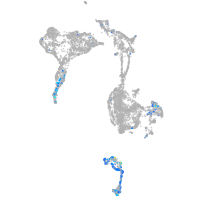
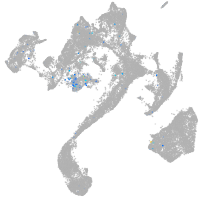
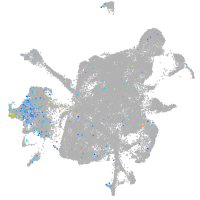
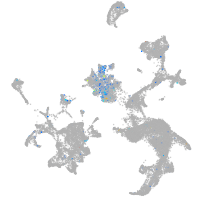
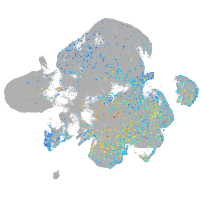
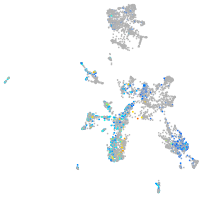
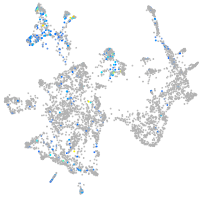
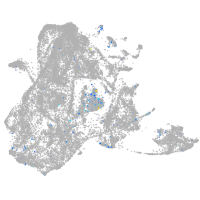
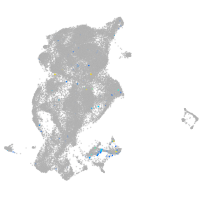
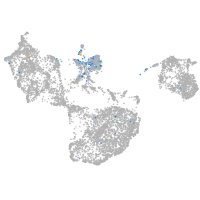
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.275 | gapdh | -0.117 |
| mir7a-1 | 0.266 | gamt | -0.112 |
| neurod1 | 0.265 | ahcy | -0.109 |
| mir375-2 | 0.257 | fbp1b | -0.107 |
| pax6b | 0.238 | gatm | -0.103 |
| si:dkey-153k10.9 | 0.235 | apoa4b.1 | -0.100 |
| fev | 0.232 | mat1a | -0.098 |
| pcsk1nl | 0.224 | apoc2 | -0.097 |
| pcsk1 | 0.223 | aldob | -0.096 |
| rnasekb | 0.223 | si:dkey-16p21.8 | -0.096 |
| nkx2.2a | 0.220 | glud1b | -0.095 |
| elovl1a | 0.219 | eno3 | -0.095 |
| zgc:101731 | 0.218 | bhmt | -0.095 |
| insm1a | 0.214 | afp4 | -0.095 |
| hepacam2 | 0.214 | gstt1a | -0.094 |
| scgn | 0.211 | aldh7a1 | -0.094 |
| calm1b | 0.209 | hspe1 | -0.093 |
| hpcal1 | 0.202 | gstr | -0.092 |
| gapdhs | 0.201 | hspd1 | -0.091 |
| tspan7b | 0.199 | apoa1b | -0.091 |
| vat1 | 0.193 | cx32.3 | -0.091 |
| gdi1 | 0.191 | suclg1 | -0.091 |
| syt1a | 0.190 | aldh6a1 | -0.090 |
| kcnj11 | 0.190 | scp2a | -0.089 |
| scn3b | 0.188 | mgst1.2 | -0.089 |
| scg2a | 0.185 | slc27a2a | -0.088 |
| myt1b | 0.185 | gcshb | -0.088 |
| egr4 | 0.183 | aqp12 | -0.088 |
| si:ch211-222k6.3 | 0.181 | apoc1 | -0.087 |
| vamp2 | 0.181 | cdo1 | -0.087 |
| syt11a | 0.181 | apobb.1 | -0.087 |
| mapk15 | 0.180 | agxtb | -0.086 |
| cacna1c | 0.178 | adka | -0.086 |
| gpc1a | 0.178 | agxta | -0.085 |
| insm1b | 0.178 | shmt1 | -0.084 |