adaptor related protein complex 3 subunit sigma 2
ZFIN 
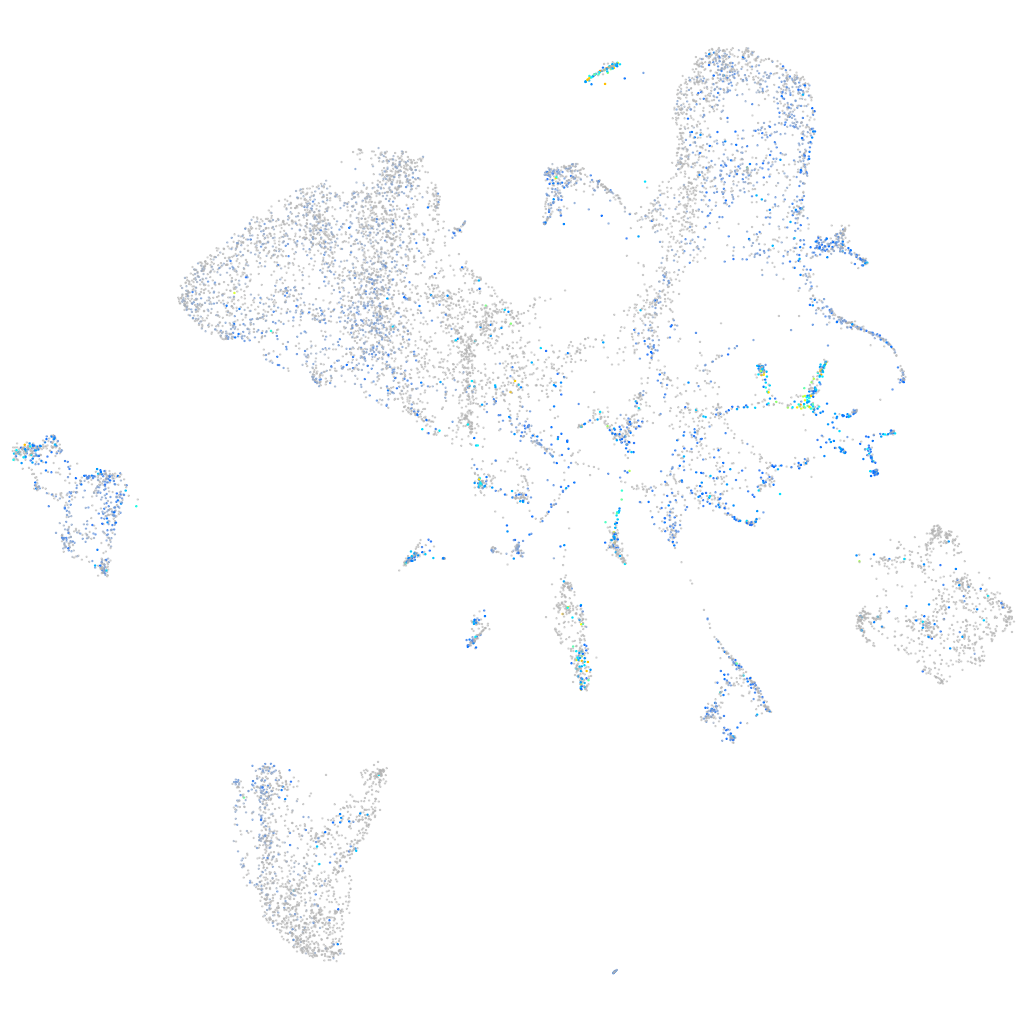
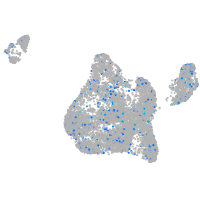
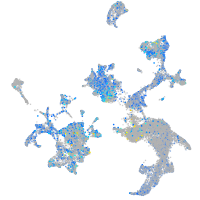
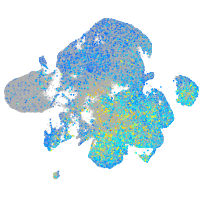
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.373 | gamt | -0.182 |
| fev | 0.344 | agxtb | -0.181 |
| calm1b | 0.319 | gapdh | -0.179 |
| egr4 | 0.317 | mat1a | -0.168 |
| neurod1 | 0.316 | pnp4b | -0.165 |
| mir375-2 | 0.313 | aqp12 | -0.163 |
| mir7a-1 | 0.312 | gatm | -0.162 |
| insm1b | 0.303 | ahcy | -0.161 |
| hepacam2 | 0.303 | nupr1b | -0.160 |
| pcsk1nl | 0.298 | bhmt | -0.160 |
| elovl1a | 0.296 | apoc1 | -0.158 |
| si:ch1073-429i10.3.1 | 0.296 | fabp10a | -0.154 |
| scgn | 0.295 | ttr | -0.152 |
| pax6b | 0.294 | gc | -0.150 |
| zgc:101731 | 0.293 | apoa1b | -0.148 |
| gapdhs | 0.291 | ces2 | -0.146 |
| calm1a | 0.284 | cx32.3 | -0.146 |
| vat1 | 0.282 | aldh6a1 | -0.145 |
| hsbp1a | 0.281 | rbp2b | -0.144 |
| dll4 | 0.277 | apoa2 | -0.142 |
| anxa4 | 0.276 | agxta | -0.141 |
| rasd4 | 0.269 | apom | -0.140 |
| nkx2.2a | 0.268 | comtd1 | -0.139 |
| c2cd4a | 0.267 | LOC110437731 | -0.139 |
| si:dkey-153k10.9 | 0.267 | hpda | -0.138 |
| id4 | 0.266 | fetub | -0.138 |
| atp6v1e1b | 0.265 | rbp4 | -0.137 |
| tmsb | 0.264 | uox | -0.137 |
| arxa | 0.264 | ttc36 | -0.136 |
| rnasekb | 0.263 | cyp2ad2 | -0.136 |
| oaz1b | 0.260 | ambp | -0.136 |
| pcsk2 | 0.259 | zgc:112265 | -0.136 |
| sat1a.2 | 0.259 | si:dkey-86l18.10 | -0.135 |
| vamp2 | 0.255 | ugp2a | -0.135 |
| rprmb | 0.254 | aldh7a1 | -0.134 |