annexin A3b
ZFIN 
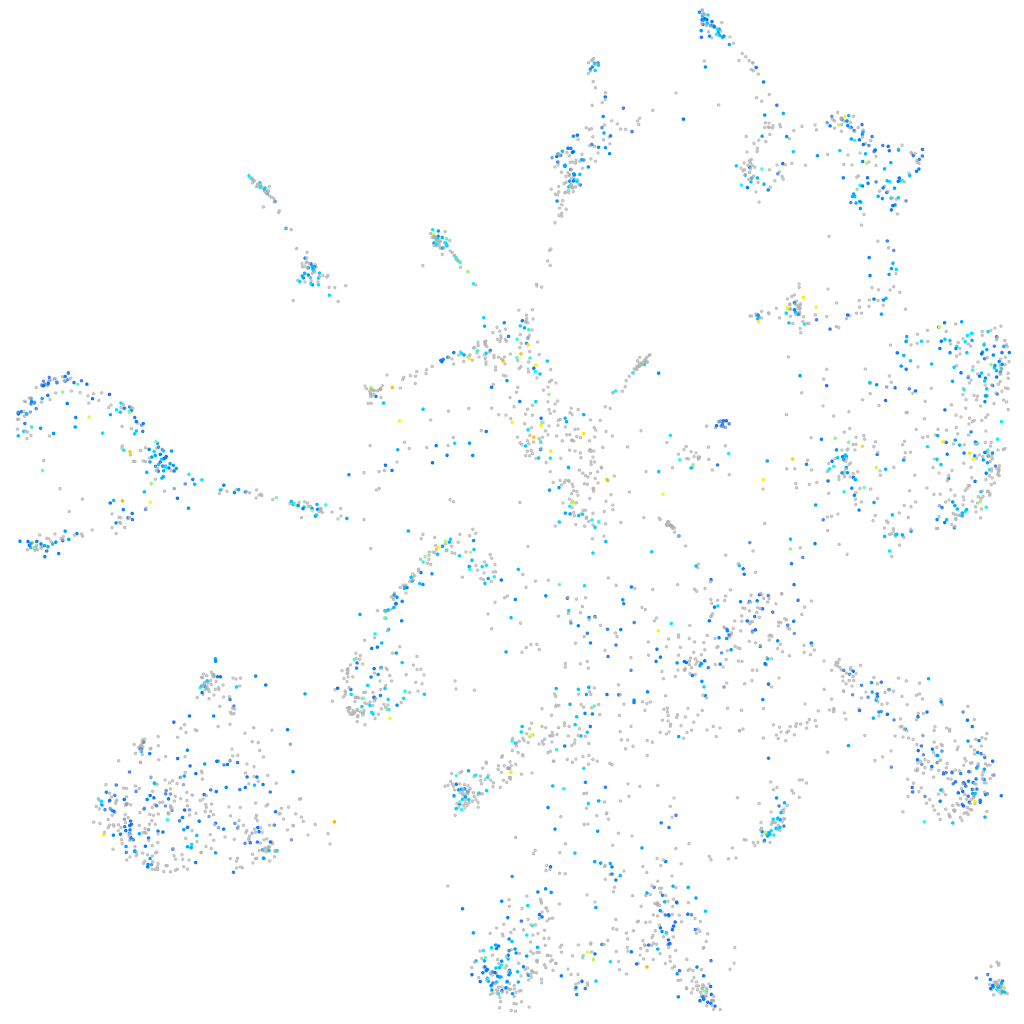
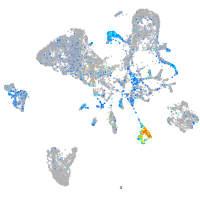
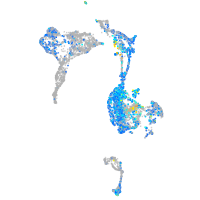
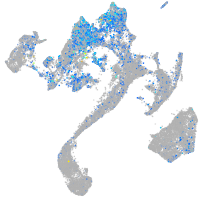
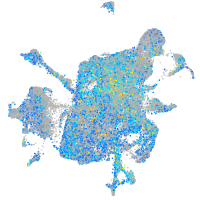
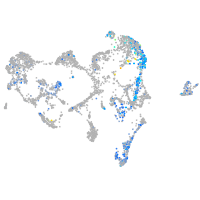
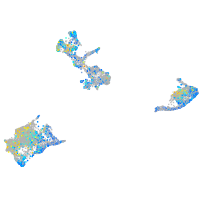
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| capns1b | 0.163 | zgc:165555.3 | -0.077 |
| krt18a.1 | 0.155 | stmn1a | -0.077 |
| zgc:153867 | 0.155 | mki67 | -0.075 |
| sparc | 0.152 | dut | -0.074 |
| myl12.1 | 0.147 | hist1h4l | -0.071 |
| capns1a | 0.145 | si:ch211-222l21.1 | -0.070 |
| s100v2 | 0.144 | zgc:153409 | -0.070 |
| arpc2 | 0.144 | si:dkey-261m9.17 | -0.066 |
| rpz5 | 0.142 | fbxo5 | -0.062 |
| arpc3 | 0.141 | NC-002333.17 | -0.062 |
| tagln2 | 0.139 | esco2 | -0.062 |
| anxa4 | 0.139 | hmgb2b | -0.061 |
| flot2a | 0.134 | ddx18 | -0.061 |
| pkma | 0.131 | NC-002333.4 | -0.061 |
| sept10 | 0.130 | sat1a.1 | -0.060 |
| anxa5b | 0.130 | elavl3 | -0.059 |
| mfap2 | 0.130 | ncapg | -0.057 |
| capzb | 0.130 | dlgap5 | -0.057 |
| actb2 | 0.130 | zgc:110425 | -0.057 |
| klf6a | 0.129 | rrm2 | -0.056 |
| cnn2 | 0.129 | lyrm7 | -0.056 |
| lasp1 | 0.129 | lig1 | -0.055 |
| cdc42l | 0.128 | tfap4 | -0.054 |
| cd63 | 0.127 | CU634008.1 | -0.054 |
| wnt11r | 0.127 | dscaml1 | -0.053 |
| pgp | 0.127 | ncapd2 | -0.053 |
| ywhaba | 0.126 | nova2 | -0.053 |
| bsg | 0.126 | si:ch211-246m6.5 | -0.053 |
| eno3 | 0.126 | cdca8 | -0.052 |
| cdc42 | 0.124 | si:ch211-113a14.18 | -0.052 |
| sri | 0.123 | mibp | -0.052 |
| ywhabb | 0.123 | im:7152348 | -0.052 |
| sdc4 | 0.122 | stmn1b | -0.051 |
| anxa11a | 0.122 | si:ch211-217k17.9 | -0.051 |
| krt8 | 0.121 | vamp2 | -0.051 |