"annexin A13, like"
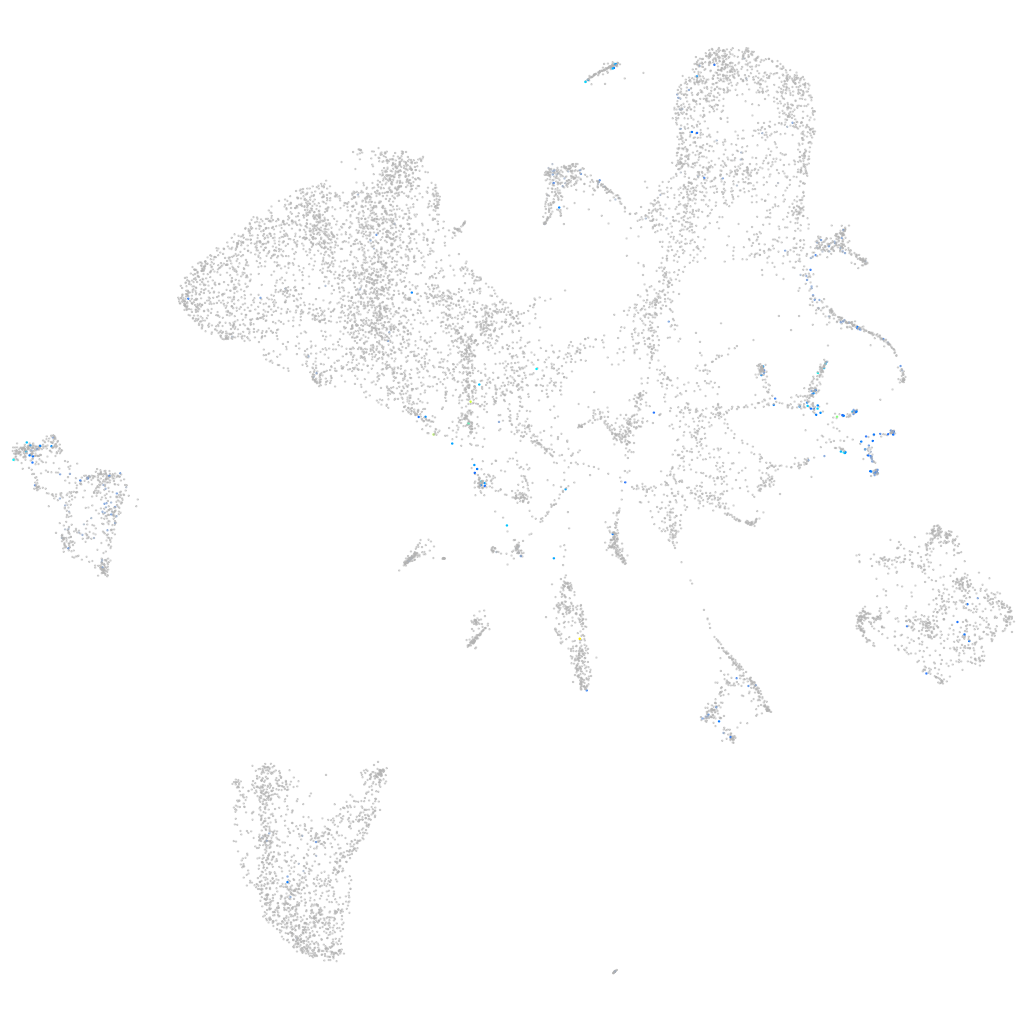
ZFIN 
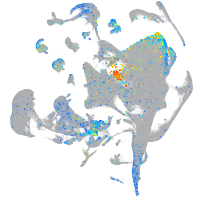
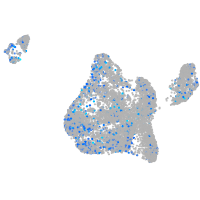
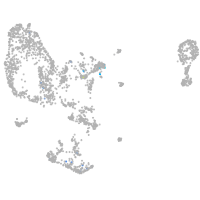
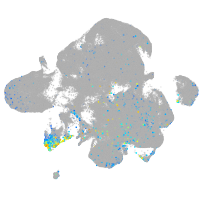
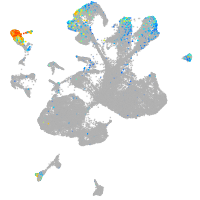
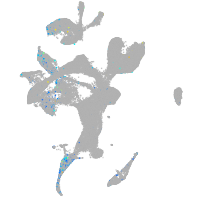
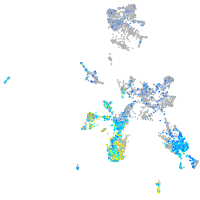
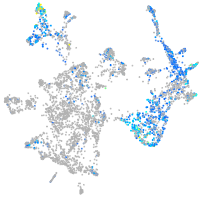
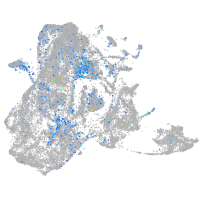
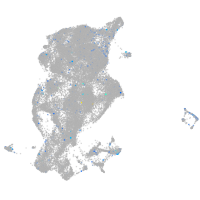
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg2a | 0.250 | gatm | -0.102 |
| scgn | 0.249 | gamt | -0.100 |
| pcsk1nl | 0.248 | gapdh | -0.095 |
| isl1 | 0.229 | ahcy | -0.094 |
| cpe | 0.224 | bhmt | -0.092 |
| syt1a | 0.223 | aqp12 | -0.090 |
| neurod1 | 0.223 | fbp1b | -0.085 |
| scg3 | 0.219 | mat1a | -0.085 |
| pcsk1 | 0.215 | nupr1b | -0.082 |
| camk1da | 0.206 | aldh6a1 | -0.082 |
| pax6b | 0.203 | cx32.3 | -0.082 |
| rnasekb | 0.202 | scp2a | -0.080 |
| trpm2 | 0.201 | apoc2 | -0.079 |
| kcnj11 | 0.200 | apoa1b | -0.078 |
| mir7a-1 | 0.199 | gcshb | -0.077 |
| pcsk2 | 0.198 | gstt1a | -0.077 |
| si:dkey-153k10.9 | 0.197 | apoa4b.1 | -0.076 |
| LOC110438591 | 0.196 | adka | -0.076 |
| si:ch73-287m6.1 | 0.192 | hspe1 | -0.076 |
| si:dkey-106n21.1 | 0.191 | agxtb | -0.075 |
| vamp2 | 0.190 | afp4 | -0.075 |
| gapdhs | 0.189 | agxta | -0.075 |
| rprmb | 0.187 | grhprb | -0.075 |
| ndufa4l2a | 0.187 | aldh7a1 | -0.075 |
| pcdh2ab5 | 0.185 | apoc1 | -0.075 |
| c2cd4a | 0.184 | glud1b | -0.075 |
| ptn | 0.183 | suclg1 | -0.075 |
| gcgb | 0.182 | pnp4b | -0.074 |
| tspan7b | 0.182 | mgst1.2 | -0.073 |
| fam49bb | 0.178 | nipsnap3a | -0.073 |
| dkk3b | 0.177 | si:dkey-16p21.8 | -0.072 |
| LOC100537384 | 0.177 | gstr | -0.072 |
| slc35g2b | 0.176 | serpinf2b | -0.071 |
| gpc1a | 0.176 | igfbp2a | -0.071 |
| egr4 | 0.174 | ttc36 | -0.071 |