ANTXR cell adhesion molecule 2a
ZFIN 
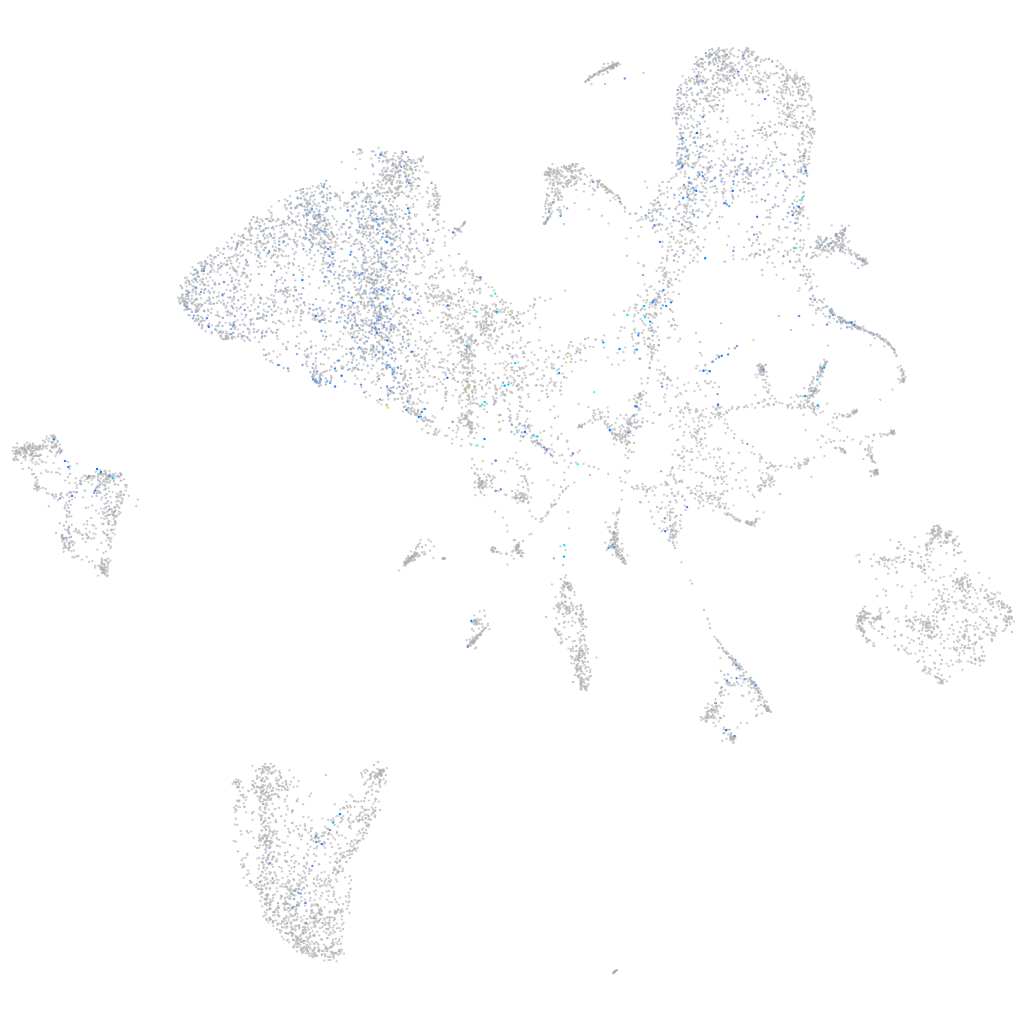
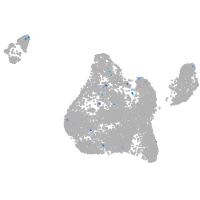
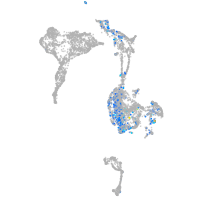
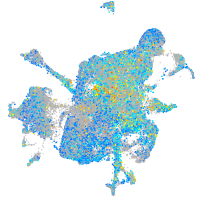
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| prdx2 | 0.144 | rtn1a | -0.092 |
| slco1d1 | 0.143 | hspb1 | -0.089 |
| LOC101885363 | 0.143 | marcksl1b | -0.079 |
| gstt1a | 0.143 | marcksb | -0.077 |
| eno3 | 0.143 | cdh6 | -0.077 |
| aldh1l1 | 0.142 | prss59.1 | -0.075 |
| ndrg1a | 0.140 | CELA1 (1 of many) | -0.074 |
| BX571762.1 | 0.140 | zgc:112160 | -0.074 |
| hspd1 | 0.139 | fep15 | -0.073 |
| hadh | 0.139 | si:ch211-195b11.3 | -0.073 |
| LOC101885014 | 0.136 | ctrl | -0.073 |
| cat | 0.136 | ctrb1 | -0.073 |
| msrb2 | 0.135 | si:ch211-152c2.3 | -0.073 |
| slc51a | 0.135 | pdia2 | -0.072 |
| mt-atp6 | 0.135 | cpa5 | -0.072 |
| mt-nd1 | 0.134 | ela2l | -0.072 |
| acadvl | 0.134 | prss59.2 | -0.072 |
| ndufb10 | 0.134 | cel.1 | -0.071 |
| cyp8b1 | 0.134 | ela2 | -0.071 |
| rdh1 | 0.134 | ela3l | -0.071 |
| slc37a4a | 0.134 | BX908782.1 | -0.071 |
| gstr | 0.133 | akap12b | -0.070 |
| si:ch73-281k2.5 | 0.133 | endou | -0.070 |
| atp5f1b | 0.132 | slc38a5b | -0.070 |
| crygm2d4 | 0.132 | apoda.2 | -0.070 |
| hadhb | 0.132 | cpb1 | -0.070 |
| vtnb | 0.132 | erp27 | -0.070 |
| scp2a | 0.132 | apoeb | -0.070 |
| ephx5 | 0.131 | si:ch211-240l19.5 | -0.070 |
| igfbp1a | 0.131 | prss1 | -0.069 |
| dpydb | 0.130 | cx43.4 | -0.069 |
| vtna | 0.130 | zgc:136461 | -0.069 |
| hadhab | 0.129 | sycn.2 | -0.069 |
| tecrb | 0.129 | amy2a | -0.069 |
| acsbg1 | 0.129 | eef1a1l1 | -0.069 |