"ankyrin repeat, family A (RFXANK-like), 2"
ZFIN 
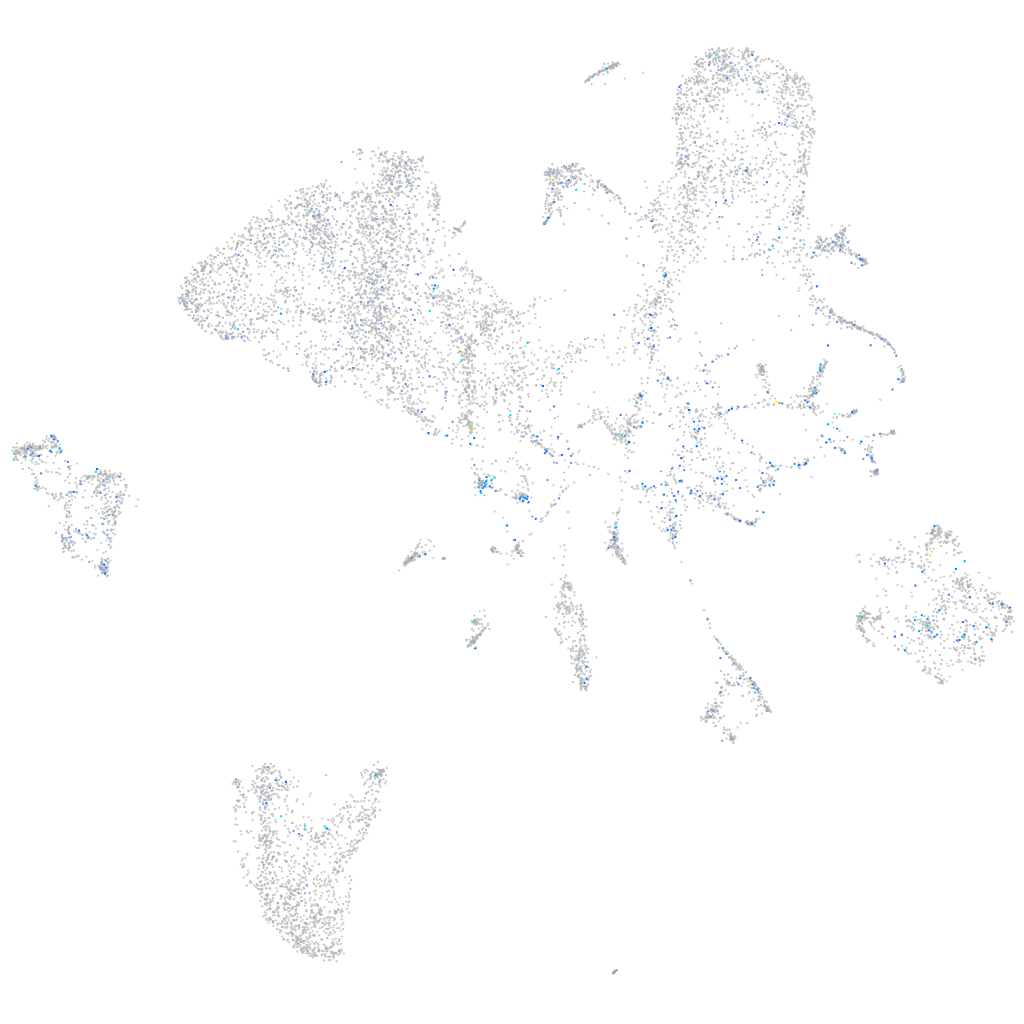
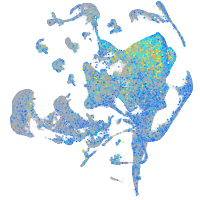
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ubc | 0.136 | gatm | -0.099 |
| si:ch1073-429i10.3.1 | 0.134 | gamt | -0.096 |
| h3f3c | 0.133 | gapdh | -0.094 |
| hnrnpa0l | 0.130 | nupr1b | -0.085 |
| h3f3d | 0.126 | ahcy | -0.083 |
| hmgn2 | 0.125 | bhmt | -0.077 |
| ywhabl | 0.125 | mat1a | -0.077 |
| wu:fb55g09 | 0.125 | pnp4b | -0.072 |
| cirbpb | 0.123 | fbp1b | -0.072 |
| si:ch211-222l21.1 | 0.123 | glud1b | -0.071 |
| hnrnpaba | 0.122 | c6ast4 | -0.071 |
| h2afvb | 0.122 | zgc:136461 | -0.071 |
| kdm6bb | 0.121 | prss59.2 | -0.071 |
| h2afx1 | 0.119 | CELA1 (1 of many) | -0.070 |
| oaz1b | 0.119 | cpa5 | -0.070 |
| ptmaa | 0.117 | ela3l | -0.070 |
| tubb4b | 0.117 | prss1 | -0.070 |
| khdrbs1a | 0.116 | apoc2 | -0.070 |
| gng3 | 0.115 | cel.2 | -0.070 |
| brk1 | 0.115 | zgc:112160 | -0.070 |
| ptmab | 0.115 | ctrb1 | -0.070 |
| drap1 | 0.115 | ela2 | -0.070 |
| tuba1c | 0.115 | prss59.1 | -0.069 |
| h3f3a | 0.114 | abat | -0.069 |
| si:ch73-1a9.3 | 0.114 | sycn.2 | -0.069 |
| ncam1a | 0.114 | apoa4b.1 | -0.069 |
| lsm7 | 0.114 | cpb1 | -0.069 |
| tmeff1b | 0.113 | agxtb | -0.069 |
| h2afva | 0.113 | zgc:92137 | -0.068 |
| si:dkey-42i9.4 | 0.113 | aqp12 | -0.068 |
| hmgb3a | 0.112 | ela2l | -0.068 |
| hmgn6 | 0.111 | si:dkey-14d8.6 | -0.068 |
| sinhcafl | 0.111 | dap | -0.067 |
| hmgb2b | 0.111 | cpa1 | -0.067 |
| hmgb1b | 0.110 | cel.1 | -0.067 |