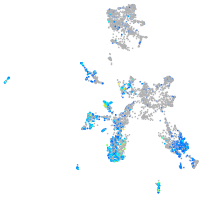
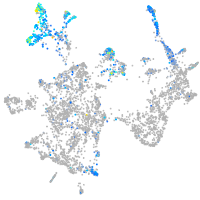
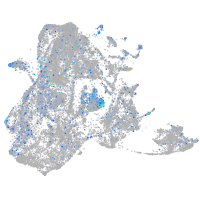
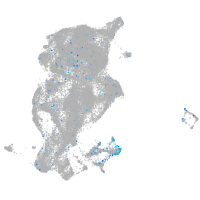
"aldolase C, fructose-bisphosphate, b"
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.459 | gapdh | -0.141 |
| syt1a | 0.439 | ahcy | -0.135 |
| pcsk1nl | 0.407 | gamt | -0.133 |
| scgn | 0.406 | aldob | -0.130 |
| pax6b | 0.397 | aldh6a1 | -0.120 |
| neurod1 | 0.395 | gatm | -0.119 |
| tspan7b | 0.392 | hdlbpa | -0.119 |
| scg2b | 0.388 | mat1a | -0.116 |
| si:dkey-153k10.9 | 0.386 | gstr | -0.116 |
| mir7a-1 | 0.373 | rps20 | -0.113 |
| eno1a | 0.373 | rpl6 | -0.112 |
| vat1 | 0.365 | cx32.3 | -0.111 |
| vamp2 | 0.363 | gstt1a | -0.108 |
| scg2a | 0.357 | nupr1b | -0.108 |
| atp6v0cb | 0.356 | aqp12 | -0.108 |
| rprmb | 0.356 | adka | -0.107 |
| pcsk1 | 0.351 | aldh9a1a.1 | -0.106 |
| cpe | 0.350 | fbp1b | -0.106 |
| atpv0e2 | 0.350 | slc16a1b | -0.104 |
| rnasekb | 0.350 | gnmt | -0.103 |
| kcnj11 | 0.343 | eno3 | -0.102 |
| rims2a | 0.337 | eef1da | -0.101 |
| sv2a | 0.337 | eif4ebp1 | -0.101 |
| isl1 | 0.336 | dhdhl | -0.101 |
| slc35g2b | 0.334 | apoa4b.1 | -0.101 |
| pcsk2 | 0.333 | bhmt | -0.101 |
| gapdhs | 0.332 | apoc1 | -0.100 |
| id4 | 0.327 | hspe1 | -0.099 |
| atp1a3a | 0.324 | nipsnap3a | -0.099 |
| gpc1a | 0.323 | aldh7a1 | -0.098 |
| insm1a | 0.319 | apoc2 | -0.097 |
| c2cd4a | 0.316 | agxtb | -0.097 |
| cplx2l | 0.316 | cpn1 | -0.095 |
| ndufa4l2a | 0.310 | cebpd | -0.095 |
| gnao1a | 0.308 | cox7a1 | -0.095 |