"aldehyde dehydrogenase 2 family member, tandem duplicate 1"
ZFIN 
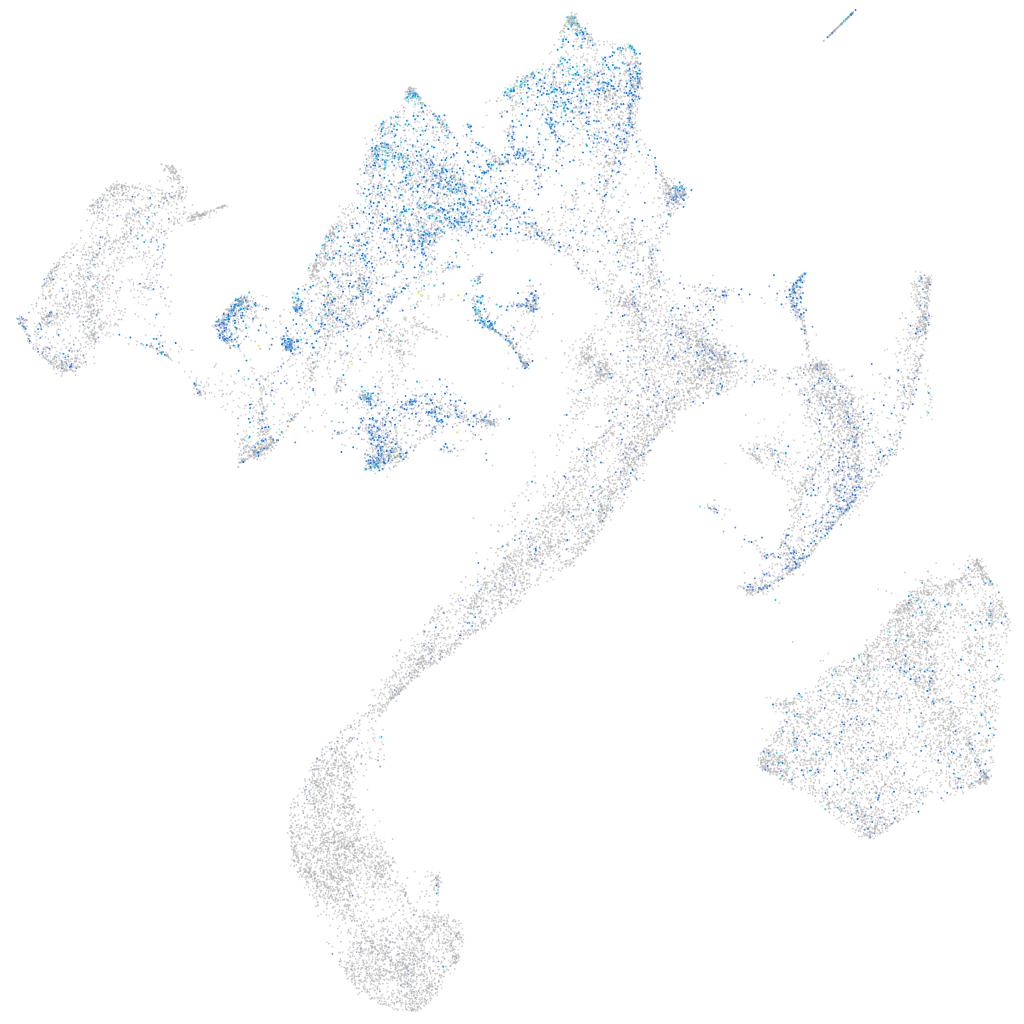
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pmp22a | 0.265 | cycsb | -0.164 |
| marcksl1a | 0.247 | hspb1 | -0.157 |
| ptmaa | 0.226 | ttn.1 | -0.145 |
| col1a2 | 0.226 | mybphb | -0.134 |
| sparc | 0.224 | aldoaa | -0.133 |
| tmsb4x | 0.224 | acta1b | -0.129 |
| foxp4 | 0.223 | tnnc2 | -0.128 |
| col1a1b | 0.221 | tpma | -0.128 |
| krt8 | 0.221 | aldoab | -0.128 |
| cd63 | 0.220 | ttn.2 | -0.126 |
| col5a1 | 0.212 | ak1 | -0.124 |
| sult1st1 | 0.208 | fxr2 | -0.123 |
| krt18b | 0.207 | CABZ01078594.1 | -0.123 |
| krt18a.1 | 0.206 | ldb3a | -0.122 |
| pdia3 | 0.205 | actn3a | -0.122 |
| postnb | 0.204 | tmem38a | -0.121 |
| tpm4a | 0.204 | neb | -0.120 |
| col1a1a | 0.203 | hsp90aa1.1 | -0.119 |
| col5a2a | 0.200 | atp2a1 | -0.119 |
| fmoda | 0.200 | gapdh | -0.118 |
| serpinf1 | 0.198 | srl | -0.118 |
| zgc:153867 | 0.197 | hhatla | -0.117 |
| mfap2 | 0.195 | si:ch73-367p23.2 | -0.116 |
| dcn | 0.194 | ldb3b | -0.116 |
| actb2 | 0.194 | zgc:92429 | -0.116 |
| anxa4 | 0.193 | ckmb | -0.115 |
| tmem176 | 0.192 | smyd1a | -0.115 |
| actb1 | 0.192 | myl1 | -0.114 |
| nme2b.1 | 0.189 | actn3b | -0.114 |
| mmp2 | 0.189 | myom1a | -0.114 |
| cd82a | 0.188 | myoz1b | -0.113 |
| ppiab | 0.187 | CABZ01072309.1 | -0.112 |
| zgc:162730 | 0.186 | rbfox1l | -0.112 |
| ppib | 0.185 | rtn2a | -0.112 |
| tmem119b | 0.185 | XLOC-001975 | -0.111 |