"aldehyde dehydrogenase 18 family, member A1"
ZFIN 
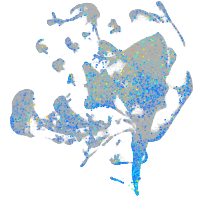
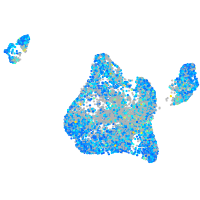
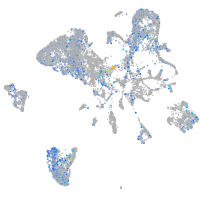
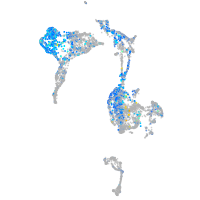
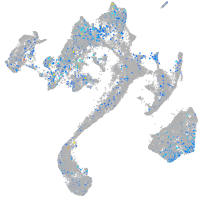
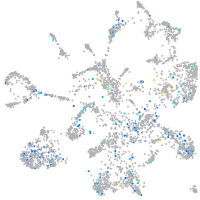
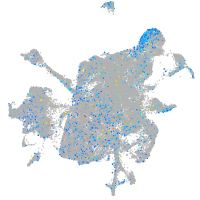
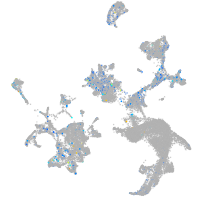
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ndufs1 | 0.285 | si:ch73-1a9.3 | -0.161 |
| si:ch211-222f23.6 | 0.248 | ptmab | -0.160 |
| sod2 | 0.238 | si:ch211-222l21.1 | -0.155 |
| dhdhl | 0.221 | hmgn6 | -0.152 |
| BX005153.2 | 0.212 | hspb1 | -0.144 |
| mctp2a | 0.208 | hmgb2a | -0.144 |
| mt-nd4 | 0.206 | h3f3d | -0.143 |
| HECTD4 | 0.204 | hnrnpaba | -0.141 |
| si:ch211-139a5.9 | 0.203 | hmgb2b | -0.140 |
| mt-nd2 | 0.202 | marcksb | -0.138 |
| glud1b | 0.200 | wu:fb97g03 | -0.137 |
| mt-atp6 | 0.197 | nucks1a | -0.134 |
| si:dkey-16p21.8 | 0.195 | hmga1a | -0.127 |
| itm2bb | 0.191 | khdrbs1a | -0.127 |
| eef1da | 0.190 | h2afvb | -0.124 |
| sdha | 0.190 | apoeb | -0.123 |
| COX3 | 0.189 | lmo2 | -0.123 |
| gstt1a | 0.189 | si:ch73-281n10.2 | -0.121 |
| aldh6a1 | 0.189 | marcksl1b | -0.120 |
| sod1 | 0.188 | apoc1 | -0.120 |
| cox6a2 | 0.187 | histh1l | -0.120 |
| glyctk | 0.186 | zgc:110425 | -0.118 |
| zgc:162944 | 0.184 | cirbpb | -0.115 |
| si:dkey-33i11.9 | 0.183 | LOC101883320 | -0.115 |
| eno3 | 0.182 | hmgn2 | -0.114 |
| fbxo15 | 0.181 | id3 | -0.114 |
| gcshb | 0.181 | rps12 | -0.112 |
| elovl1b | 0.180 | acin1a | -0.111 |
| slc26a6l | 0.180 | lin28a | -0.111 |
| phyhd1 | 0.179 | hmgn3 | -0.108 |
| mt-co1 | 0.179 | syncrip | -0.107 |
| cx32.3 | 0.178 | rbm8a | -0.107 |
| CR381686.5 | 0.178 | tardbp | -0.106 |
| psap | 0.178 | eef1a1l1 | -0.106 |
| atp5f1b | 0.178 | hnrnpabb | -0.106 |