"aldo-keto reductase family 1, member B1 (aldose reductase) [Source:ZFIN;Acc:ZDB-GENE-040625-7]"
ZFIN 
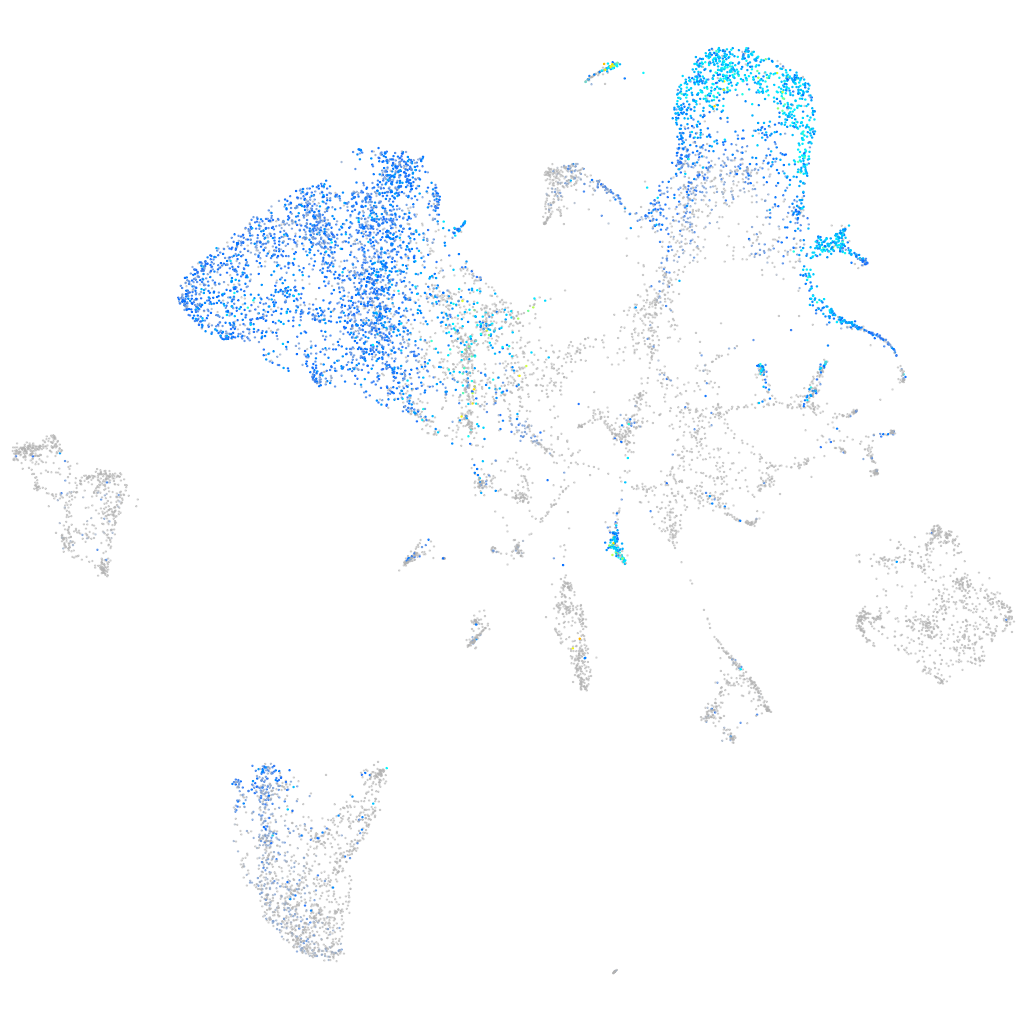
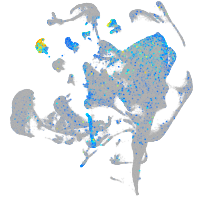
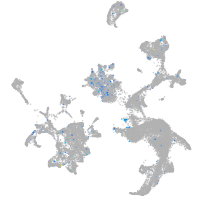
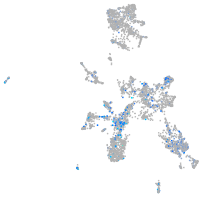
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cyp3a65 | 0.581 | slc38a5b | -0.301 |
| glud1b | 0.562 | si:ch211-222l21.1 | -0.292 |
| sod1 | 0.557 | hmgb3a | -0.287 |
| agmo | 0.550 | hmgb1b | -0.278 |
| rdh1 | 0.549 | marcksl1b | -0.276 |
| dhrs9 | 0.543 | marcksb | -0.276 |
| srd5a2a | 0.536 | si:ch73-1a9.3 | -0.273 |
| faxdc2 | 0.532 | hmgb2a | -0.269 |
| gpx4a | 0.529 | nucks1a | -0.269 |
| si:ch211-133l5.7 | 0.521 | glulb | -0.269 |
| abcc2 | 0.520 | ptmab | -0.267 |
| gapdh | 0.511 | si:ch73-281n10.2 | -0.267 |
| idh1 | 0.510 | cirbpa | -0.262 |
| scp2a | 0.509 | h2afvb | -0.261 |
| si:ch211-107o10.3 | 0.508 | cirbpb | -0.259 |
| apoa4b.1 | 0.506 | cx43.4 | -0.255 |
| eno3 | 0.506 | stmn1a | -0.255 |
| mdh1aa | 0.506 | bcam | -0.254 |
| fbp1b | 0.505 | anp32b | -0.254 |
| prdx2 | 0.503 | qkia | -0.254 |
| sult2st2 | 0.501 | syncrip | -0.254 |
| tpi1b | 0.499 | seta | -0.254 |
| ugt1a7 | 0.496 | anp32a | -0.253 |
| zanl | 0.494 | id1 | -0.252 |
| tm4sf5 | 0.493 | epcam | -0.249 |
| ada | 0.492 | hmgn2 | -0.247 |
| sdr16c5b | 0.491 | hmgb2b | -0.246 |
| clic5b | 0.491 | hmgn6 | -0.246 |
| pck1 | 0.490 | hnrnpa0a | -0.243 |
| dhrs1 | 0.489 | khdrbs1a | -0.242 |
| gstr | 0.487 | hnrnpa1a | -0.241 |
| chia.2 | 0.483 | tpm4a | -0.239 |
| abcg2a | 0.483 | hnrnpaba | -0.238 |
| AL831745.1 | 0.482 | hmga1a | -0.238 |
| atp5mc1 | 0.478 | cbx5 | -0.238 |