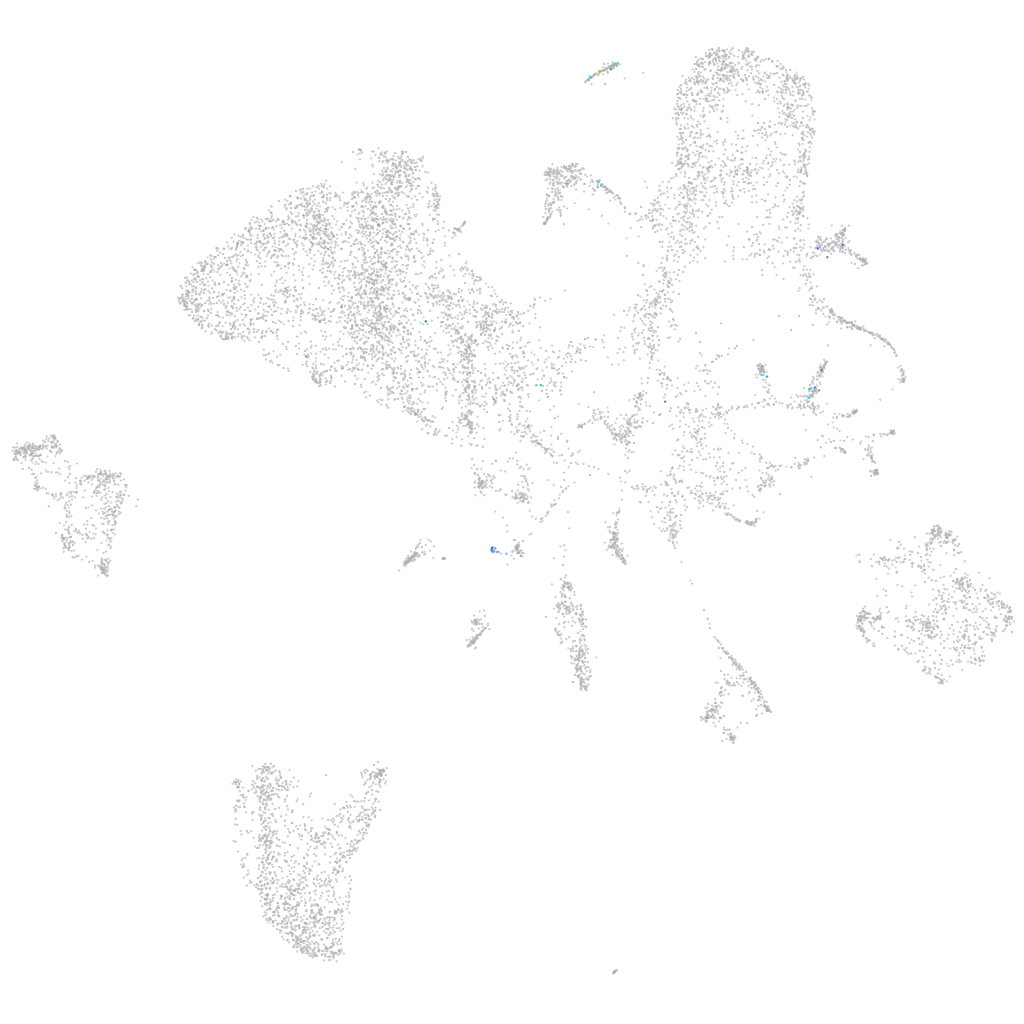
adenylate kinase 9
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 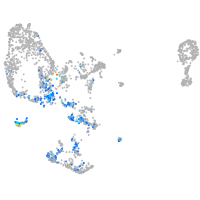 |
neural |
spinal cord / glia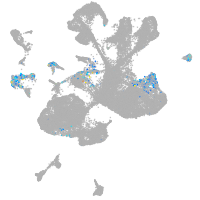 |
eye |
taste / olfactory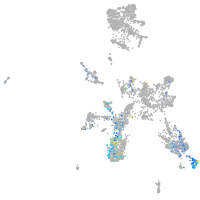 |
otic / lateral line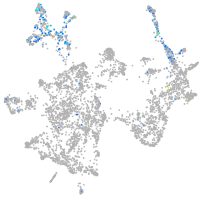 |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| lrguk | 0.270 | npm1a | -0.035 |
| zbbx | 0.252 | ssr1 | -0.034 |
| cfap126 | 0.227 | rps12 | -0.034 |
| spag16 | 0.221 | fabp3 | -0.033 |
| CABZ01085594.1 | 0.213 | rpl7l1 | -0.032 |
| lrrc51 | 0.211 | sec61b | -0.031 |
| crocc2 | 0.210 | hdlbpa | -0.031 |
| gabra2a | 0.209 | apoeb | -0.031 |
| fam155a | 0.207 | tram1 | -0.031 |
| LOC103912072 | 0.207 | ahcy | -0.030 |
| tbata | 0.198 | ybx1 | -0.030 |
| dlec1 | 0.182 | fbl | -0.030 |
| nme5 | 0.182 | pes | -0.029 |
| CABZ01079081.1 | 0.182 | dap | -0.028 |
| mapk15 | 0.179 | ssr2 | -0.028 |
| si:ch1073-166e24.4 | 0.178 | cnbpa | -0.028 |
| spata18 | 0.177 | eif4ebp3l | -0.028 |
| LOC103911624 | 0.171 | eif4ebp1 | -0.028 |
| capsla | 0.169 | gatm | -0.028 |
| pih1d2 | 0.168 | etf1b | -0.028 |
| or137-3 | 0.167 | impdh1b | -0.027 |
| cfap57 | 0.167 | ssr4 | -0.027 |
| alkal2b | 0.164 | cpb1 | -0.027 |
| CR855320.1 | 0.159 | apex1 | -0.027 |
| CR855277.2 | 0.159 | gspt1l | -0.026 |
| gmnc | 0.156 | zgc:112160 | -0.026 |
| enkur | 0.155 | nip7 | -0.026 |
| cfap52 | 0.155 | ela2 | -0.026 |
| avpr1aa | 0.154 | tars | -0.026 |
| CU571064.1 | 0.154 | fep15 | -0.026 |
| pmfbp1 | 0.154 | erp27 | -0.026 |
| c18h15orf65 | 0.153 | pdia2 | -0.026 |
| gabrr2b | 0.152 | cgref1 | -0.026 |
| tspan3b | 0.151 | apoda.2 | -0.026 |
| ccdc151 | 0.150 | c6ast4 | -0.025 |