aminoacyl tRNA synthetase complex interacting multifunctional protein 2
ZFIN 
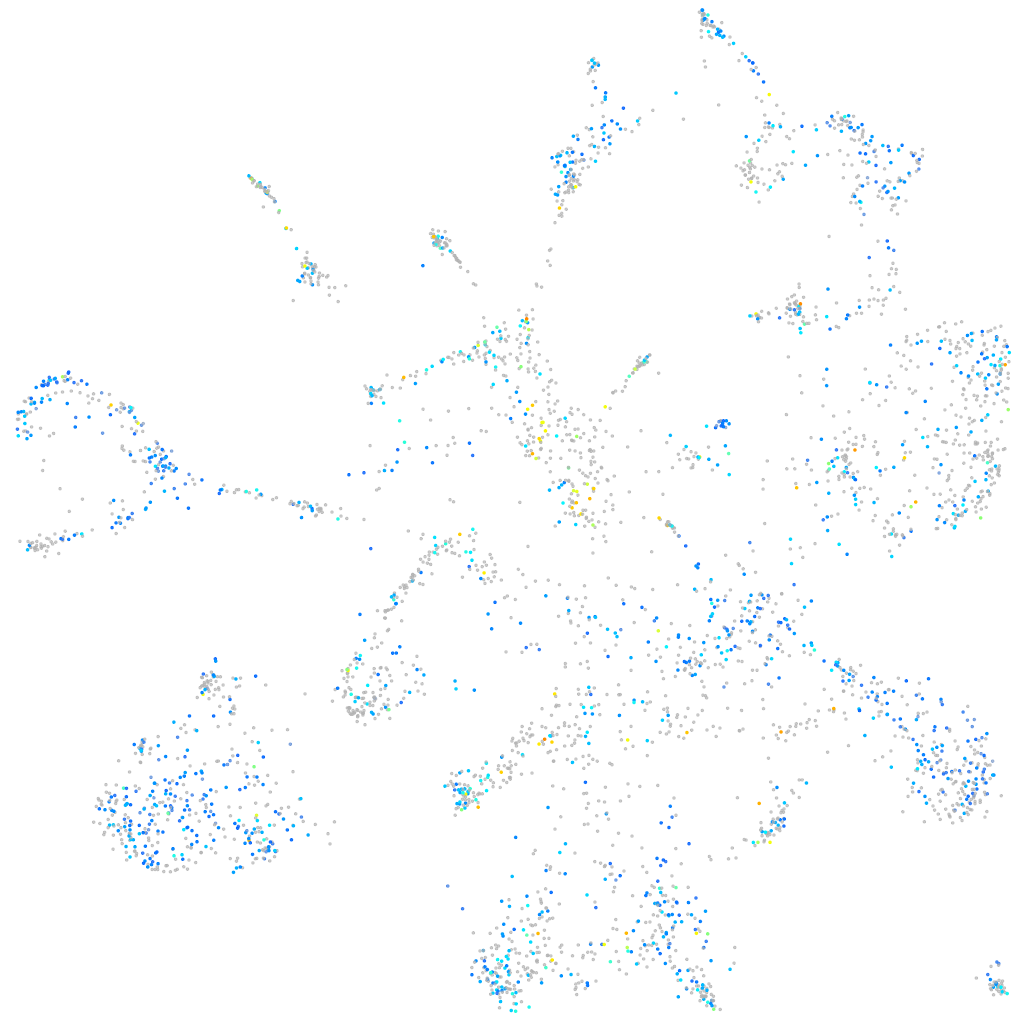
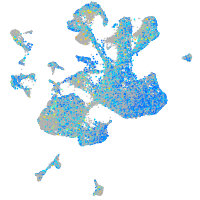
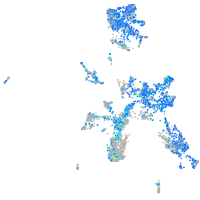
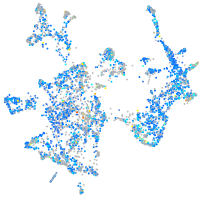
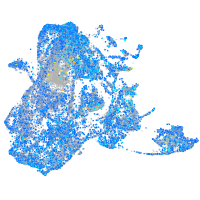
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cct5 | 0.113 | rhag | -0.070 |
| mdh2 | 0.110 | tpma | -0.057 |
| ran | 0.109 | myhz1.1 | -0.057 |
| ldhba | 0.108 | mylpfa | -0.055 |
| rpl19 | 0.106 | NC-002333.4 | -0.053 |
| mibp2 | 0.102 | CABZ01072614.1 | -0.052 |
| ybx1 | 0.101 | hbae1.1 | -0.051 |
| rpl8 | 0.099 | zgc:158463 | -0.049 |
| eef1g | 0.099 | shroom2a | -0.049 |
| atp5mc3b | 0.099 | hbbe1.1 | -0.049 |
| hsp90ab1 | 0.098 | txn | -0.048 |
| cstf2 | 0.098 | myl1 | -0.048 |
| naca | 0.097 | tcf7l1b | -0.047 |
| arf5 | 0.097 | actc1b | -0.046 |
| cct7 | 0.096 | col18a1a | -0.046 |
| ndufb10 | 0.096 | lamc1 | -0.045 |
| cct2 | 0.095 | pvalb1 | -0.044 |
| atp5f1b | 0.094 | LOC110439372 | -0.044 |
| ptges3a | 0.094 | COX3 | -0.044 |
| rpl13 | 0.094 | itga8 | -0.043 |
| hnrnpaba | 0.093 | serpina1l | -0.043 |
| rps2 | 0.093 | pvalb2 | -0.043 |
| bcas2 | 0.093 | kif26aa | -0.042 |
| bysl | 0.092 | ttn.2 | -0.040 |
| tomm20b | 0.092 | ahr2 | -0.040 |
| rplp0 | 0.092 | fbxl3b | -0.040 |
| psmb6 | 0.091 | hbae3 | -0.039 |
| setb | 0.090 | dennd4c | -0.039 |
| smdt1b | 0.090 | tnnt3b | -0.038 |
| vdac1 | 0.090 | mylpfb | -0.038 |
| slc25a3b | 0.090 | lama5 | -0.038 |
| COX5B | 0.089 | nt5e | -0.038 |
| vdac2 | 0.089 | aqp1a.1 | -0.037 |
| polr2f | 0.088 | si:ch211-285j22.3 | -0.037 |
| tmed2 | 0.088 | edc3 | -0.037 |